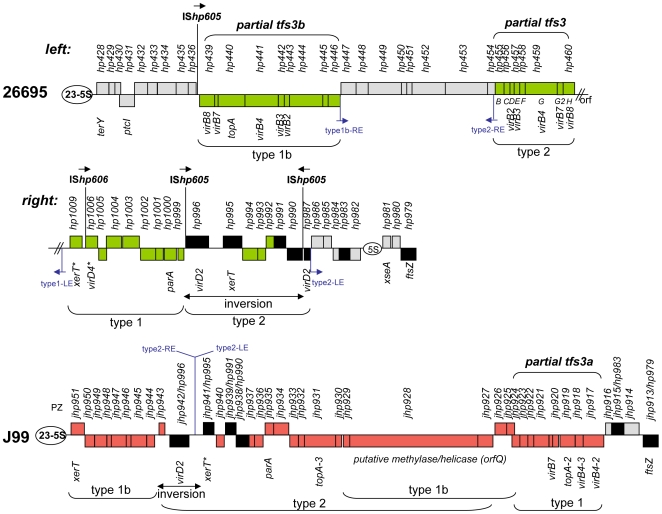
Figure 3. Plasticity zones of reference strains 26695 and J99 ([8], [18], GenBank Accessions AE000511 and AE001439).
Color coding: boxes in green, TnPZ orfs found in 26695 but not J99; in red, TnPZ orfs found in J99 but not 26695; in black, orfs found in both strains; in grey, orfs located between ftsZ gene and 5S, 23S rRNA gene pair that are judged to be not of TnPZ origin. TnPZ left and right ends are designated LE and RE, with arrows pointing outward. Strain 26695. This segment is inferred to have resulted from multiple TnPZ insertions, and deletions that left only remnants of original full-length elements. It was also split in two by a large chromosomal inversion (breakpoint indicated by //). This breakpoint is associated with a type 1 TnPZ, suggesting that it had mediated the inversion. Strain J99. This plasticity zone (jhp0917-jhp0951) is located between the ftsZ (jhp0913) gene and the 5S,23S rRNA gene pair, and is composed of fragments from type 1 (jhp0917-jhp0924), 1b (jhp0944-jhp0951; jhp0925, jhp0926 potentially part of jhp0927-jhp0929 (orfQ) as well); and type 2 (jhp0943-jhp0930 and potentially part of orfQ as well) TnPZs. A lack of clustering of orfQ gene sequences according to TnPZ types, however, precluded more precise identification of the boundary between type 1b and type 2 sequences. jhp0942 and flanking regions (0.9 kb next to 5′-end, 1.15 kb next to 3′-end, which also contains a short jhp0943 orf), are inverted relative to nearby genes. One inversion breakpoint coincides with juxtaposed left and right ends of a type 2 TnPZ. jhp0941 (labeled xerT) is a composite orf in which the first 114 xerT codons have been replaced by 90 codons of orfB of tfs3. These interpretations are provisional, however, because the J99 genomic DNA that had been provided to the genome sequencing team contained a tfs3 gene cluster and a second copy of orfQ [24], which were not included in the reported genome sequence [8], and the possibility of additional errors has not been assessed.