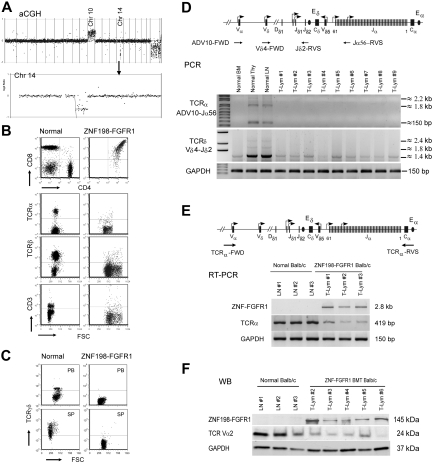
Figure 4.
Frequent loss of TCRα and TCRδ in lymph tissues in ZNF198-FGFR1 mice. (A) A representative whole genome aCGH profile from a mouse lymphoma showed 14qC2 segmental loss as well as gain of Chr:10. (B) Flow cytometric analysis revealed that lymphocytes from a serial transplanted mouse are DP with negative TCRα, TCRβ, and CD3e. (C) Both peripheral WBCs and splenocytes from leukemic mice are TCRγδ negative using the anti-TCRγδ. (D) Genomic DNA PCR analysis, using primers located within the TCRα ADV10 variable segment (forward) and joint region 56 (reverse, top panel), was used to analyze the TCRα rearrangement. A forward primer located in the TCRδ Vδ4 segment and a reverse primer located in the Jδ2 region (top panel) to amplify the TCRδ rearrangement were used. Both PCR analyses suggested that TCRα and TCRδ rearrangements were absent in the peripheral lymph tissues from ZNF198-FGFR1 mice. (E) Semiquantitative RT-PCR, using a forward primer located in TCRα variable region and a reverse primer located in the constant, C, region (top panel), showed that TCRα transcription levels were significantly reduced in T lymphomas compared with normal mesenteric LNs. (F) Western blot analysis showed positive expression of the ZNF198-FGFR1 fusion protein, but low expression of TCR Vα2 in lymphoid tissues of disease mice.