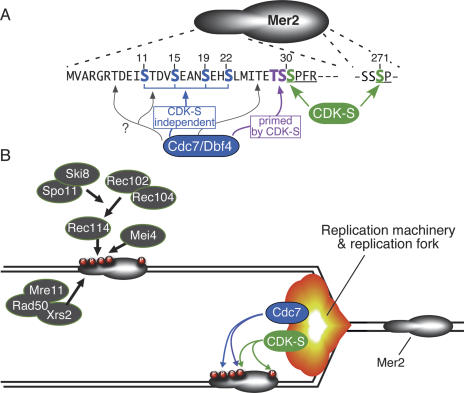
Figure 2.
CDK-S and Cdc7/Dbf4 regulate DSB formation via phosphorylation of Mer2. (A) Phosphorylation sites on Mer2. Amino acid sequence is shown for the N-terminal region (residues 1–33) and a phosphorylation site near the C terminus (residues 269–272). Green indicates residues phosphorylated by CDK-S (Ser30 and Ser271). Two types of Cdc7/Dbf4 target are shown: Blue indicates serine residues phosphorylated by Cdc7 independently of CDK-S, and magenta indicates residues phosphorylated by Cdc7 only after CDK-S has primed the sites by phosphorylating the adjacent serine. Three additional threonine residues (gray arrows) are present in this region and are also potential Cdc7 targets. See the text for further details. (B) Model for coupling of replication with DSB formation. Mer2 is proposed to be preferentially phosphorylated in the wake of replication fork passage, perhaps through physical association of CDK-S and/or Cdc7/Dbf4 with components of the replication machinery. Phosphorylated Mer2 is then able to recruit other DSB proteins, including Spo11 itself. See the text for further details.