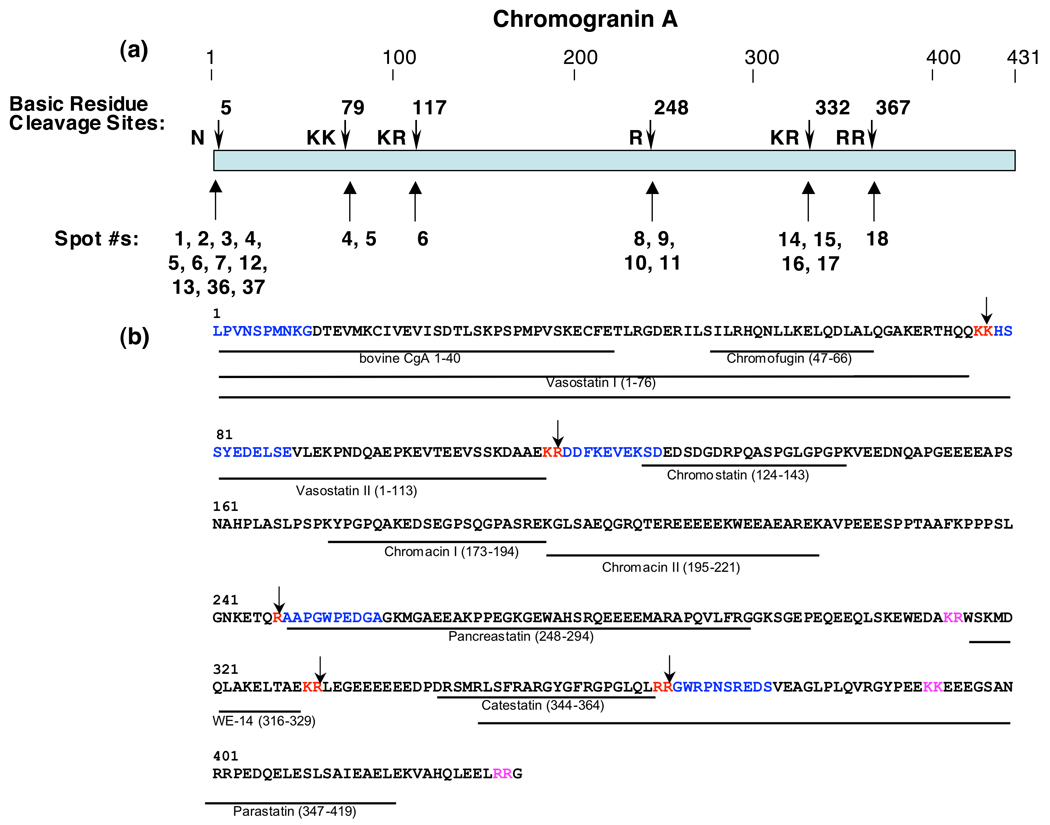
Figure 2. Cleavage sites of full-length CgA that generate proteolytic fragments in chromaffin granules.
a. Cleavage sites utilized to generate NH2-termini of CgA proteins in chromaffin granules. Cleavage sites of CgA representing NH2-termini of CgA fragments are shown by the arrows. The protein spots (numbered as in figure 1) whose NH2-termini correspond to the indicated cleavage sites are shown below the cleavage sites (shown by arrows).
b. Cleavage sites and biologically active peptides within full-length CgA. Cleavage sites utilized to generate CgA fragments in chromaffin granules are illustrated by the arrows at dibasic or monobasic residue sites shown in red font. NH2-terminal sequence determined by Edman degradation of CgA fragments are illustrated in blue color font, within the full-length CgA primary sequence; dibasic residues at cleavage sites are shown in red font. Biologically active peptides derived from CgA are shown as lines with peptide names under the bovine CgA primary sequence (NCBI Protein Database accession number P05059 (30)); cleavage sites predicted to generate these active fragments that were not identified in this study are shown in pink font.