Abstract
We provide a demonstration in humans of the principle of pharmacometabonomics by showing a clear connection between an individual's metabolic phenotype, in the form of a predose urinary metabolite profile, and the metabolic fate of a standard dose of the widely used analgesic acetaminophen. Predose and postdose urinary metabolite profiles were determined by 1H NMR spectroscopy. The predose spectra were statistically analyzed in relation to drug metabolite excretion to detect predose biomarkers of drug fate and a human-gut microbiome cometabolite predictor was identified. Thus, we found that individuals having high predose urinary levels of p-cresol sulfate had low postdose urinary ratios of acetaminophen sulfate to acetaminophen glucuronide. We conclude that, in individuals with high bacterially mediated p-cresol generation, competitive O-sulfonation of p-cresol reduces the effective systemic capacity to sulfonate acetaminophen. Given that acetaminophen is such a widely used and seemingly well-understood drug, this finding provides a clear demonstration of the immense potential and power of the pharmacometabonomic approach. However, we expect many other sulfonation reactions to be similarly affected by competition with p-cresol and our finding also has important implications for certain diseases as well as for the variable responses induced by many different drugs and xenobiotics. We propose that assessing the effects of microbiome activity should be an integral part of pharmaceutical development and of personalized health care. Furthermore, we envisage that gut bacterial populations might be deliberately manipulated to improve drug efficacy and to reduce adverse drug reactions.
Keywords: acetaminophen, p-cresol, bacteria, sulfate, glucuronide
The effects of drug treatments can vary greatly between different individuals, and pharmacogenomics has been widely advocated as a potential means of personalizing human drug treatments to increase drug efficacy and to decrease adverse reactions (1–6). However, environmental factors (such as nutritional status, gut bacterial activities, age, disease, and other drug use) are also important determinants of individual metabolic phenotypes, which modulate drug metabolism, efficacy, and toxicity. Such environmental complications, which may also alter gene expression, will tend to limit the usefulness of predictions of drug-induced responses that are based only on genomic differences (7, 8). For instance, for many classes of compound, enzyme induction state, which is environmentally determined, influences drug metabolism and toxicity and this is not captured in genomic data. Recognizing this important limitation of pharmacogenomics, a different approach to personalized drug treatment has recently been proposed wherein predose metabolite profiling would instead be used to predict a subject's responses to potential drug interventions (9). This “pharmacometabonomic” approach has a number of major advantages, which include the ready availability and relative ease of analysis of biofluids, such as urine and blood plasma, as well as the fact that the derived metabolite profiles are sensitive to both genomic and environmental influences affecting metabolism. A further crucial advantage and feature of this metabonomic approach is its openness to finding unexpected biomarkers and biomarker combinations, as multiple analytes are quantified simultaneously without prespecification of what those analytes should be (10). The only factors limiting which analytes are detected are the nature of the sample that is analyzed and the analytical platform used. Thus, pharmacometabonomic modeling need not be limited by prior understanding or hypothesis. However, despite much support and enthusiasm for the concept (9, 11–13), there has, until now, to the best of our knowledge, been no convincing pharmacometabonomic demonstration in humans.
To test the feasibility of applying the pharmacometabonomic approach to man, we chose as our example the well-known analgesic and antipyretic drug acetaminophen (N-acetyl-p-aminophenol; known as paracetamol in Europe). Acetaminophen is one of the most widely used nonprescription medicines in the world and its toxicology and metabolism have been extensively investigated over many years (14–20). However, we will show here that, even for this most familiar drug, pharmacometabonomic analysis will yield significantly increased understanding of its metabolic behavior in humans. These findings have considerable implications for personalized drug treatment in general and lead to new and testable hypotheses for a number of diseases.
Acetaminophen was chosen to exemplify the pharmacometabonomic principle for a variety of reasons, which included its common usage and its low toxicity at therapeutic doses, which was necessary to establish an ethically approved clinical trial. It is also predominantly and relatively rapidly eliminated in the urine (15–19). Thus, by collecting postdose urine samples we could study the manner of its excretion by each subject, such excretion being known to show considerable intersubject variation (20). Precisely how a particular drug is metabolized and excreted by each individual can have a major influence on its safety and efficacy. Thus, for instance, greater or lesser production of a toxic metabolite might be expected to influence the extent of an adverse effect, whereas the rate of removal of the pharmacologically active compound might be expected to influence the extent and duration of the desired pharmacological action. With this in mind, the aim of the present study was to determine whether metabonomic analysis of predose human urine would allow prediction of some aspect of acetaminophen metabolism or excretion at the individual subject level and, thereby, provide a proof-of-principle for the feasibility of pharmacometabonomics in humans.
Our ethically approved study (SI Text) was based on 99 healthy male volunteers who were all nonsmokers between 18 and 64 years old with a condition of their participation being that they had not taken any drugs in the preceding week. Each participant provided a predose urine sample and then, after taking a standard therapeutic dose of acetaminophen (two 500-mg tablets), each was requested to collect all of his postdose urine over 2 consecutive 3-h periods (0–3 h and 3–6 h after dosing). All of the samples were analyzed by 600 MHz 1H NMR spectroscopy with the predose spectra providing profiles of the detectable, naturally occurring (endogenous) metabolites and the postdose spectra providing profiles of the acetaminophen-related compounds superimposed on an endogenous metabolite “background.” From these NMR spectra and the known urine volumes, the urinary acetaminophen-related compounds excreted by each subject over each postdose collection period were quantified as acetaminophen sulfate (S), acetaminophen glucuronide (G), and “other,” with the “other” components expected to be mainly the parent, cysteine conjugate, and N-acetylcysteine conjugate (17–19). We then searched for components of intersubject variation in the predose spectra that would correlate with intersubject variation in the postdose data. However, from the outset, prediction of the S/G ratio was of particular interest because this ratio is known to show extensive interindividual variation and is assumed to be indicative of the relative extent to which acetaminophen is metabolized via 2 major phase 2 conjugative processes (O-sulfonation and glucuronidation) that impact on the metabolism of many different drugs (20, 21). Additionally, we expected that the S/G ratio would be less susceptible to any sample collection errors than the absolute amounts of metabolites excreted.
Results and Discussion
The Urinary Excretion of Acetaminophen and Its Metabolites.
We found that ≈30% of the 1-g dose was recovered (as any acetaminophen-related compound) in each of the 2 postdose urine collection periods (0–3 h and 3–6 h), this being consistent with the findings of an earlier report (15) where the urinary excretion of acetaminophen, S and G was studied at 1.5-h intervals after a 12 mg/kg dose. Furthermore, we found that, in terms of moles, the combined excretion of S and G typically accounted for ≈85% of the total amount of acetaminophen-related compounds recovered in each collection period, this being consistent with the 24-h urinary recoveries from 111 Caucasians given a 1.5-g dose (20). Representative predose and postdose spectra are shown in Fig. 1.
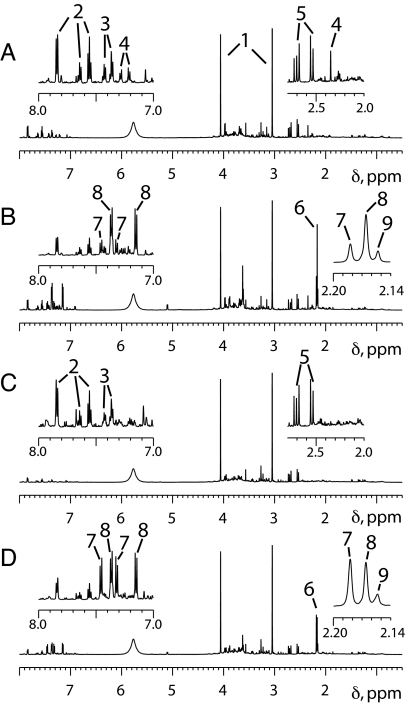
Fig. 1.
Representative 1H NMR spectra of urine samples provided before and after taking 1 g of acetaminophen. (A) The δ 8.0–0.5 region of the predose urine spectrum for a subject whose urine contained a relatively high level of p-cresol sulfate. (B) The corresponding 0–3 h postdose urine spectrum, which shows a relatively low ratio of acetaminophen sulfate to acetaminophen glucuronide. (C) The δ 8.0–0.5 region of the predose urine spectrum for a subject whose urine did not contain a high level of p-cresol sulfate. (D) The corresponding 0–3 h postdose spectrum, which shows a relatively high ratio of acetaminophen sulfate to acetaminophen glucuronide. To facilitate their comparison, all these spectra were processed in the same way, without resolution enhancement and with a digital filter used to minimize the residual water features, which would otherwise be observed at ≈δ 4.7. Furthermore, each spectrum has been scaled so that the creatinine methylene peak at ≈δ 4.06 is just on scale (with the result that the corresponding creatinine methyl peak at ≈δ 3.05 is off scale in each case). The Insets, which are expansions of selected spectral regions, are scaled to fill the available space. Key to numbered peaks: 1, creatinine; 2, hippurate; 3, phenylacetylglutamine; 4, p-cresol sulfate; 5, citrate; 6, cluster of N-acetyl groups from acetaminophen-related compounds; 7, acetaminophen sulfate; 8, acetaminophen glucuronide; 9, other acetaminophen-related compounds.
The average S/G ratios for the 2 postdose collections were found to be 0.71 (0–3 h) and 0.53 (3–6 h), respectively, with the S/G value for each individual subject always being lower in the 3–6 h collection than in the 0–3 h collection, and this change in the S/G ratios being consistent with what has been reported for the early excretion of a 20-mg/kg dose (17). Furthermore, there was a clear correlation between the S/G 0–3 h and S/G 3–6 h data (r = 0.927). However, a plot of these data indicated an outlier and it was judged, from this and from an unusually low excretion of acetaminophen-related metabolites, that this subject had not fully collected his 0–3 h sample. With this subject excluded, the correlation between S/G 0–3 h and S/G 3–6 h was slightly improved (r = 0.948). A moderate (17%) decrease in average S excretion was observed between the 2 collection periods along with a largely compensating increase in average G excretion. Having excluded the 0–3 h data for the 1 subject who appeared not to have collected his 0–3 h urine properly, the correlation coefficients between the observed S/G ratios and the amount of S excreted were found to be 0.510 (0–3 h) and 0.835 (3–6 h), respectively. Likewise, the correlation coefficients between S/G and the amount of G excreted were found to be −0.738 (0–3 h) and −0.803 (3–6 h), respectively. Precautionary checks showed that the amounts of S and G excreted, and the S/G ratio, were not related to the age of the subjects or to their body mass or to the order in which the samples were analyzed (Table S1).
Examination of the Predose Spectral Profiles.
Our initial way of searching for relationships between the predose and postdose data was by means of the PLS (Projection to Latent Structure)-based pattern recognition methods that are typically used in metabonomic studies (22) and which had proved highly effective in our earlier animal-based work (9). However, in the present case, the PLS-based approach was relatively unproductive (SI Text) and, subsequently, we made a detailed visual comparison of creatinine-normalized predose spectra for subjects at the 2 ends of the S/G ratio distribution (25 subjects at each end). With this revised and relatively simple approach, we found 2 potentially discriminatory predose metabolites, later identified as the microbial cometabolites p-cresol sulfate (PCS) and phenylacetylglutamine (PAG), with higher levels of these metabolites being visually associated with lower S/G values (Figs. 1 and 2). These findings were supported by principal-components analyses (PCA) focused on the aromatic region (δ 9.1–6.9) of the predose NMR spectra (SI Text and Fig. S1), and no other components of the predose spectra were found to have such clear discriminatory potential in regard to the S/G ratios observed. On closer inspection, the predose urinary levels of PCS and PAG were found to be broadly correlated (r = 0.75), which, in retrospect, was unsurprising because there is a significant degree of commonality in their origins before the final sulfate and glutamine conjugations; thus, p-cresol and phenylacetic acid are known to be produced from tyrosine and phenylalanine, respectively, with these conversions being largely analogous and dependent on the action of colonic bacteria (23) (Fig. 3). However, our further analysis, using integrated predose spectral band intensities and numerical single variable discovery procedures, showed that, of all the individual spectral components, only PCS was likely to provide statistically significant discrimination with respect to S/G (SI Text).
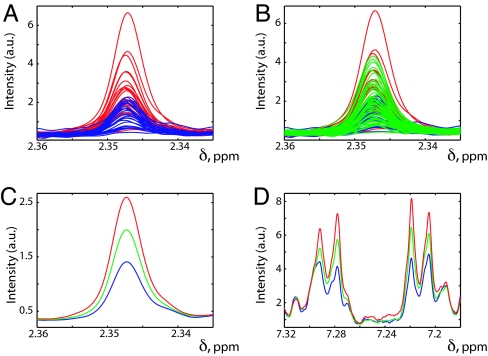
Fig. 2.
Selected regions of 1H NMR spectra obtained from predose urine samples with color-coding according to postdose behavior. All of the plots were produced in MATLAB with each individual NMR spectrum being normalized to constant creatinine. (A) An expansion of the δ 2.335–2.360 spectral region, which contains the methyl signal from p-cresol sulfate (PCS), with the individual spectra for the 25 subjects giving the highest postdose 0–3 h S/G ratios shown in blue and superimposed on the individual spectra for the 25 subjects giving the lowest postdose 0–3 h S/G ratios shown in red. (B) The same plot as A but with the further addition of the corresponding data for the other 49 subjects (shown in green). (C) The same spectral region and the average spectra for the 3 different groups, with the same color coding. (D) The same average predose spectra, with the same color coding, over the region of δ 7.18–7.32, which contains the PCS aromatic signals (the pair of pseudo “doublets” centered at ≈δ 7.21 and at ≈δ 7.29). In all plots “a.u.” designates arbitrary units. In plots A and B, some spectra are obscured by the subsequently superimposed spectra.
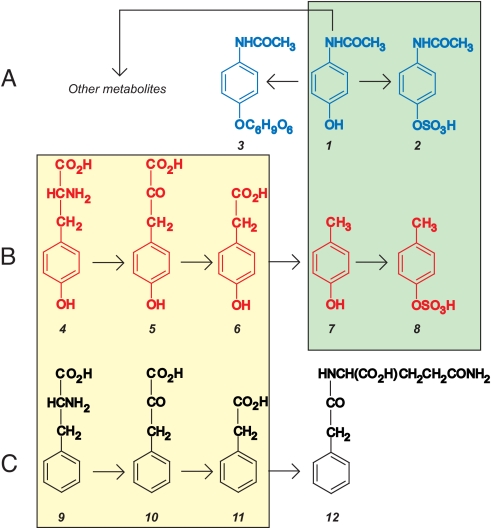
Fig. 3.
Relevant metabolic pathways. (A) The hydroxyl group of acetaminophen (1) may be sulfonated to produce acetaminophen sulfate (2) or glucuronidated to produce acetaminophen glucuronide (3). (B) Stepwise production of p-cresol sulfate (8) from tyrosine (4) (23). The green box highlights the highly analogous and potentially competitive sulfonation of acetaminophen and p-cresol (25, 26). (C) Stepwise production of phenylacetylglutamine (12) from phenylalanine (9) (23) with the yellow box highlighting similarities with the metabolism of tyrosine. Key to compounds: 1, acetaminophen; 2, acetaminophen sulfate; 3, acetaminophen glucuronide; 4, tyrosine; 5, 4-hydroxyphenylpyruvic acid; 6, 4-hydroxyphenylacetic acid; 7, p-cresol; 8, p-cresol sulfate; 9, phenylalanine; 10, phenylpyruvic acid; 11, phenylacetic acid; 12, phenylacetylglutamine. In the body, compounds 2 and 8 would normally be expected to exist as ROSO3− rather than as ROSO3H, where R designates the remainder of each molecule.
Further Analyses Focused on PCS.
To get the best possible measure of the level of PCS relative to creatinine, which, for this study, was a suitable internal reference compound for the urinary quantitation (SI Text), selected peaks in the predose NMR spectra were then integrated after local baseline correction. The peaks chosen for integration were the PCS methyl singlet at ≈δ 2.35 and the creatinine methylene singlet at ≈δ 4.06 and the relevant integral ratios [designated I.R., where I.R. = (integral of PCS methyl)/(integral of creatinine methylene)] obtained for each of the 99 subjects are presented in Fig. 4, where these predose data are plotted against the corresponding S/G ratios for the 2 postdose collections. From inspection of Fig. 4, it is readily apparent that a high predose level of PCS (I.R. > 0.06) is associated with a low S/G ratio postdose and use of the Mann–Whitney U test in conjunction with an appropriate Bonferroni correction (100) (SI Text) confirmed the statistical significance of the distribution of the high PCS group (25 subjects) with respect to the S/G ratios obtained in each postdose collection. The Bonferroni correction was applied to counter the multiple hypothesis testing that results from the multivariate nature of metabonomic data (24). With a Bonferroni correction of 100, the P value for 95% confidence becomes 0.05/100 = 5 × 10−4 and the P values obtained from the Mann–Whitney tests were 1.0 × 10−4 (for S/G 0–3 h) and 1.2 × 10−4 (for S/G 3–6 h).
Fig. 4.
The observed relationship between the predose urinary ratio of p-cresol sulfate (PCS) to creatinine and the postdose urinary ratio of the major acetaminophen metabolites: acetaminophen sulfate (S) and acetaminophen glucuronide (G). (A) The predose urinary PCS/creatinine integral ratio for each subject plotted against the corresponding urinary S/G ratio obtained in the 0–3 h postdose collection. (B) The corresponding plot for the 3–6 h postdose collection. I.R. designates the integral ratio of the peaks at ≈δ 2.35 and at ≈δ 4.06 in the 1H NMR spectrum recorded from the predose urine sample. For equimolar PCS and creatinine, I.R. would be expected to be approximately 1.5 because of the number of protons contributing to each signal. No subjects were excluded from either plot.
In the preceding analysis of the full data set (all subjects included), we found a high predose level of PCS to be associated with a low S/G ratio postdose and, clearly, that postdose ratio could be affected by variation in the amounts of both S and G excreted. However, because the conversion of p-cresol to PCS is analogous to the conversion of acetaminophen to S (Fig. 3), the observed connection to predose PCS strongly suggests that the relevant controlling factor is the amount of S excreted. Furthermore, with lower S/G ratios being observed in the 3–6 h collection than in the 0–3 h collection, it appears that 1 g of acetaminophen represents a substantial challenge to the sulfonation capacity of the subjects studied. The extent to which any compound undergoes sulfonation can potentially be limited both by the availability of the sulfonate donor, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), and by the characteristics and availability of the relevant sulfotransferase enzyme (25). Thus, in the present context, it is particularly notable that p-cresol and acetaminophen are both substrates for the same human cytosolic sulfotransferase, SULT1A1 (26), and can, therefore, compete for enzyme binding sites as well as for PAPS. Furthermore, in contrast to what has been reported for rats (27), recent literature suggests that p-cresol is almost entirely converted to PCS in humans (28–30). Thus, we envisaged that an individual's capacity to sulfonate acetaminophen will be reduced by ongoing presentation of endogenous p-cresol, and the potential competitive significance of the p-cresol challenge was confirmed by calculation (SI Text). On the basis of this hypothesis, we examined, with the full data set, the postdose excretion of both S and G and found that, in the 3–6 h collection, the high predose PCS subjects (I.R. > 0.06) were clearly associated with lower S excretion (P = 2.3 × 10−5 with 95% confidence at P = 5 × 10−4 after Bonferroni correction). Furthermore, a similar and statistically significant (P = 1.6 × 10−4) relationship was found when the S excretion values for this collection period were first corrected to unit body mass. Thus, these data are fully consistent with the hypothesis that substantial predose production of endogenous p-cresol can reduce an individual's ability to sulfonate acetaminophen by acting as a competitive substrate (Fig. 3). As regards the site of this competitive sulfonation, the colonic mucosa is known to have significant sulfonation capacity (25, 31) and could potentially convert colonically produced p-cresol to PCS. However, with acetaminophen being rapidly absorbed from the small intestine and with the liver being regarded as the principal site of its metabolism (17), we envisage that some colonically produced p-cresol may escape further colonic modification and be sulfonated in the liver rather than in the gastrointestinal tract.
Influence of Experimental Variables.
To check for experimental variables that might be influencing these data, the distribution of the 25 high predose PCS (I.R. > 0.06) subjects was also investigated with respect to analytical run order and with respect to the data obtained for subject age, height, body mass, and body mass index (BMI), but no association was found. However, it was noticeable that the 10 subjects showing the highest predose PCS levels (I.R. > 0.09) tended to be older (P = 0.01) and shorter (P = 0.02) individuals who would, in general, be expected to have less muscle mass and to excrete less creatinine. However, the first of these findings also suggests that aging might lead to increased p-cresol production, and this could potentially be caused by age-related changes in the nature of the gut bacteria (32). Furthermore, whereas, in the present study, we found no clear evidence of a relationship between the S/G ratio and age, an age-related decrease in acetaminophen sulfonation has been observed in male rats (33). To further check the basis of our findings we also reexamined the data after first excluding all those subjects where there was any known or suspected noncompliance with the study protocol [e.g., where a sensitive analysis of a subject's predose urine sample suggested some prior use of acetaminophen or where there was evidence of recent alcohol consumption (SI Text)]. With the remaining 78 subjects, the graphical relationship between predose PCS and the S/G ratio was maintained [the P values for the distribution of the high PCS subjects (I.R. > 0.06) with respect to S/G 0–3 h and S/G 3–6 being 7.4 × 10−4 and 9.9 × 10−4, respectively] and statistical significance was still achieved for the distribution of the high predose PCS subjects with respect to the absolute amount of S excreted during the 3–6 h collection (P = 3.4 × 10−4). We, therefore, conclude that the perceived predose to postdose connection is real.
Potential Biomedical Significance.
Although the potential significance of gut bacteria in relation to human metabolism, disease, and drug-induced reactions is becoming increasingly well recognized (34–41), we have not seen any previous report of the present finding, which could be of considerable importance if it can be proven to hold for the wider human population or for specific subsets of that population.
We envisage that, by depleting hepatic sulfonation capacity, continual exposure to colonically produced p-cresol would leave the liver more vulnerable to acetaminophen-induced damage and that markedly increased p-cresol production could potentially explain the reported association between fasting and an increased likelihood of hepatotoxicity from acetaminophen (42, 43). However, in principle, sustained prior exposure to colonically produced p-cresol could also potentially increase acetaminophen hepatotoxicity by other means, such as by enzyme induction or glutathione depletion (44–46), and preliminary data (SI Text) suggest that high p-cresol exposure might lead to a more generalized impairment of sulfur-dependent reactive metabolite detoxification, with PAPS depletion possibly leading to depletion of both taurine and glutathione. However, it remains to be investigated if our present finding has any significance for adverse reactions to acetaminophen. Instead, the wider and more obvious significance of our finding lies in its potential consequences for sulfonation reactions in general and in suggesting a potentially causal link between certain diseases and the gut bacteria.
Many different compounds are substrates for sulfotransferase-catalyzed sulfonation, which, by making them more hydrophilic, has a major role in modifying the physical properties of both small and large molecules. Thus, sulfonation facilitates the excretion of many compounds and is crucial to the structure and properties of macromolecules such as chondroitin sulfate (a component of cartilage). Notably, many drugs and/or their hydroxylated metabolites are phase II conjugated via sulfonation. Among several other important functions, sulfonation is also known to have a role in modulating the action of hormones and neurotransmitters and appears to be especially important during early human development (21, 25, 26, 47–51). There are various human sulfotransferases (both cytosolic and membrane-bound), but human cytosolic sulfotransferase SULT1A1, which acts on acetaminophen, has a broad substrate range and is one of the most important sulfotransferases for xenobiotic sulfonation as well as acting on several endogenous substrates (26, 51). Additionally, a key feature of sulfonation in higher organisms is that all such reactions use PAPS as the universal sulfonate donor. Thus, we might reasonably expect that, by competing for PAPS or for one or more sulfotransferases, the flux of p-cresol through the system will affect the sulfonation of a wide range of drugs and endogenous compounds, thereby influencing normal bodily processes as well as drug metabolic fate, efficacy, and toxicity. However, given what is known about gut bacterial production of p-cresol from protein residues (23, 43, 52–54) (Fig. 3), with Clostridium difficile being one of a number of p-cresol producers (53, 54), our present results show that environmental factors can exert a dominant influence on the extent to which a compound becomes sulfonated in the human body. Thus, we would expect that, by altering the amount of p-cresol produced, variation in either the diet or the gut bacteria could potentially exert a major influence on drug-induced responses or diseases where sulfonation has an important role.
In its role as a hypotensive agent, minoxidil provides one example of a drug where sulfonation is considered to be important in producing the desired pharmacological effect (55). However, sulfonation is not always beneficial, and tamoxifen, which is used in treating breast cancer, provides an example of a drug where sulfonation has been suggested to be important for the development of an associated adverse reaction, namely an increased incidence of endometrial cancer. Thus, it has been suggested that tamoxifen-DNA adducts are formed via O-sulfonation (56). As a further example, sulfonation phenotype could potentially influence both the efficacy and side-effects of apomorphine, for which sulfonation is the major metabolic pathway in humans (57). As regards known associations with disease, hyperactivity in children provides one example of a condition that has been associated with increased p-cresol levels and where the involvement of dietary factors is also suspected (58). Furthermore, an increased urinary level of PCS has been associated with the progression of multiple sclerosis (59, 60). Additionally, various other diseases (Parkinson disease, motor neuron disease, rheumatoid arthritis, and childhood autism) have been associated with a reduction in the S/G ratios obtained after acetaminophen dosing (61–63), which leads us to tentatively suggest, on the basis of our current findings, that excessive gut bacterial p-cresol production might also have some relevance to their etiology, with further circumstantial evidence coming from additional associations with gastrointestinal abnormalities (64–67). However, it should be clearly recognized that these various associations do not prove a causal role for p-cresol in respect of these diseases and also that p-cresol can exert a variety of effects (27–30, 43, 45, 46, 68–73) such as blocking the conversion of the neurotransmitter dopamine to noradrenaline (68). Therefore, as far as we are aware, any involvement of p-cresol in respect to these diseases remains to be proven as well as the exact nature of any such involvement. However, one general hypothesis would be that where the diet or the profile of the gut bacteria is altered in favor of p-cresol production, impaired sulfonation and other effects can result such that, depending on a subject's individual characteristics and state of development, a variety of consequences would be expected.
Conclusions and Future Prospects.
In the population studied, in this, our first pharmacometabonomic study in humans, we have found a clear association between an individual's predose urinary metabolite profile and the postdose urinary fate of acetaminophen. Further investigation will be required to determine the extent to which this association holds for the wider human population, but it is encouraging that, in hindsight, it makes such clear biochemical sense. Thus, our findings strongly suggest that a person's capacity for acetaminophen sulfonation can be significantly reduced by competitive p-cresol sulfonation, with p-cresol known to be produced from protein-derived tyrosine in reactions involving gut bacteria. Given the range of substances for which sulfonation is important, this finding suggests a means by which the gut bacteria might influence both drug-induced responses and disease development. Furthermore, given that acetaminophen is so widely used and has been extensively studied over many years, our findings provide a remarkable demonstration of the power and potential of the pharmacometabonomic approach, which we hope will eventually be used to improve drug treatment outcomes. With a view to the future practicality of this exciting approach, it is also encouraging that the present result was obtained without the potential advantage of a standard diet and using only “snapshot” predose urine samples. Furthermore, we envisage that rapidly growing recognition of the multiple metabolic interactions between humans and their gut symbionts, and the potential significance of the latter in regard to disease, drug efficacy, and adverse drug reactions, will lead to a revolution in the way that drugs are developed. We also envisage that, in certain cases, gut bacteria will be the principal target of drug action (41), and that in some other cases, gut bacteria will be manipulated by some prior or accompanying treatment in order to improve drug treatment outcomes.
Materials and Methods
Full details are provided in the SI Text. The study volunteers were nominally healthy men between 18 and 64 years old. Urine samples were prepared for NMR analysis by mixing 440 μL of urine with 220 μL of phosphate buffer (pH ≈7.4 to which sodium azide had been added as an antibacterial preservative), and the mixture was centrifuged to remove suspended particles. 550 μL of “clear” buffered urine was transferred to a sample vial and 55 μL of a TSP/D2O solution was added to give a final TSP concentration of 1 mM. TSP (sodium 3-trimethylsilyl-[2,2,3,3-2H4]-1-propionate) is a chemical shift reference compound used in the NMR experiment and the D2O provided a field/frequency lock for the NMR spectrometer. The 1H NMR spectra of the prepared urine samples were acquired at 303 K on a Bruker Avance 600 NMR spectrometer equipped with a flow probe and operated by means of the Xwinnmr software (all from Bruker Biospin). The “noesypresat” pulse sequence was used to suppress the water signal and to acquire the data, and the spectral acquisitions were automated by using the Iconnmr software and a BEST sample changer (both Bruker Biospin). The acquired data were processed using Xwinnmr. 1-Hz line-broadening was applied to the predose 1H NMR spectra by means of an exponential multiplication of the free induction decay signal and these time-domain data were Fourier-transformed to frequency-domain spectra with a single zero-filling and with a digital filter used to reduce the size of the residual water signal. The resulting spectra were manually phased to give an even baseline around the NMR signals, and the baseline of each spectrum was manually adjusted to zero intensity by using a straight-line baseline correction algorithm. The chemical shift scale was set by assigning the value of δ 0 to the signal from the added reference compound (TSP). The postdose 1H NMR spectra were processed similarly, using 0.3-Hz line broadening, and subsequently with resolution enhancement, and a measure approximating to the mole ratio of acetaminophen sulfate (S) to acetaminophen glucuronide (G) was determined by integration of the respective resolution-enhanced N-acetyl peaks. For each subject and collection period, relative measures of the amounts of S and G excreted were also determined, by reference to the added TSP and the mass of urine collected. Except where stated, these excretion values were not adjusted to excretion per unit of body mass. Subsequently, the predose spectra were loaded into MATLAB using in-house software and normalized to a constant integral for the δ 4.07–4.05 region, which encompasses the creatinine methylene singlet. Average group spectra were then calculated in MATLAB and various spectral plots were made and examined. After integrating each predose spectrum over consecutive 0.04-ppm spectral segments and then normalizing these integrals to a constant value for the integral for the δ 4.07–4.05 region, principal-components analysis of the integrals for the δ 9.1–6.9 region was performed in Pirouette 3.11 (from Infometrix) using mean-centered variable scaling. The PCA was also repeated after normalization to a constant integral for the δ 3.07–3.03 region, which encompasses the creatinine methyl singlet (Fig. S1). Local baseline correction and integration of selected peaks in the predose spectra was performed in Xwinnmr. Statistical significance of abstracted data was assessed by using the Mann–Whitney U test. See also Tables S1–S4.
Supplementary Material
Acknowledgments.
We thank Prof. R. L. Smith of Imperial College London for his advice regarding the design of the study; the staff of the Pfizer Research Centre, Canterbury, Kent, U.K., for conducting the dosing and sample collection phase; the study volunteers for their participation; Dr. C. Legido-Quigley of King's College London for conducting supporting UPLC-MS analyses; Dr. Bernard North (Imperial College Statistical Advisory Service) for statistical advice and analysis; Dr. O. Cloarec (Royal Holloway, University of London) for the use of MATLAB routines that he developed while at Imperial College London and for associated guidance; Mr. J. T. M. Pearce of Imperial College London for MATLAB support; Prof. H. Tang (now of Wuhan University, People's Republic of China) and Dr. O. Beckonert of Imperial College London for NMR support; and Dr. D. A. Parker of Imperial College London for providing data that assisted in the identification of PAG. We acknowledge the prior contributions of Dr. H. Antti (University of Umeä, Sweden) and Ms. R. Walley (Pfizer, U.K.) in identifying the potential relationship between predose creatinine and the total excretion of acetaminophen-related compounds over the first postdose collection period (SI Text). The authors thank Pfizer Global R & D for funding this work and T.A.C. thanks Pfizer for personal financial support. J. K. N. is a member of the Imperial College MRC-HPA Center for Environment and Health.
Footnotes
Conflict of interest statement: T.A.C., J.C.L., J.R.E., and J.K.N. are inventors on a relevant patent application from which financial gain might be derived. T.A.C., J.C.L., and J.K.N. might also benefit financially from the future placement of related analytical or research contracts.
See Commentary on page 14187.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904489106/DCSupplemental.
References
- 1.Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Johnson JA. Pharmacogenomics: The inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- 3.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 4.Pagliarulo V, Datar RH, Cote RJ. Role of genetic and expression profiling in pharmacogenomics: The changing face of patient management. Curr Issues Mol Biol. 2002;4:101–110. [PubMed] [Google Scholar]
- 5.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 6.Marsh S, McLeod HL. Pharmacogenomics: From bedside to clinical practice. Hum Mol Genet. 2006;15:R89–R93. doi: 10.1093/hmg/ddl087. [DOI] [PubMed] [Google Scholar]
- 7.Nebert DW, Jorge-Nebert L, Vesell ES. Pharmacogenomics and “individualized drug therapy”: High expectations and disappointing achievements. Am J Pharmacogenomics. 2003;3:361–370. doi: 10.2165/00129785-200303060-00002. [DOI] [PubMed] [Google Scholar]
- 8.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: Past lessons, future directions. Drug Metab Rev. 2008;40:187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton TA, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 10.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomic technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 11.Nebert DW, Vesell ES. Can personalized drug therapy be achieved? A closer look at pharmaco-metabonomics. Trends Pharmacol Sci. 2006;27:580–586. doi: 10.1016/j.tips.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Li H, et al. Pharmacometabonomic phenotyping reveals different responses to xenobiotic intervention in rats. J Proteome Res. 2007;6:1364–1370. doi: 10.1021/pr060513q. [DOI] [PubMed] [Google Scholar]
- 13.Ala-Korpela M. Potential role of body fluid 1H NMR metabonomics as a prognostic and diagnostic tool. Exp Rev Mol Diagn. 2007;7:761–773. doi: 10.1586/14737159.7.6.761. [DOI] [PubMed] [Google Scholar]
- 14.Newson RB, Shaheen SO, Chinn S, Burney PGJ. Paracetamol sales and atopic disease in children and adults: An ecological analysis. Eur Respir J. 2000;16:817–823. doi: 10.1183/09031936.00.16581700. [DOI] [PubMed] [Google Scholar]
- 15.Cummings AJ, King ML, Martin BK. A kinetic study of drug elimination: The excretion of paracetamol and its metabolites in man. Br J Pharmacol Chemother. 1967;29:150–157. doi: 10.1111/j.1476-5381.1967.tb01948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clements JA, Heading RC, Nimmo WS, Prescott LF. Kinetics of acetaminophen absorption and gastric emptying in man. Clin Pharmacol Ther. 1978;24:420–431. doi: 10.1002/cpt1978244420. [DOI] [PubMed] [Google Scholar]
- 17.Prescott LF. Kinetics and metabolism of paracetamol and phenacetin. Br J Clin Pharmacol. 1980;10:291S–298S. doi: 10.1111/j.1365-2125.1980.tb01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrest JAH, Clements JA, Prescott LF. Clinical pharmacokinetics of paracetamol. Clin Pharmacokinet. 1982;7:93–107. doi: 10.2165/00003088-198207020-00001. [DOI] [PubMed] [Google Scholar]
- 19.Lau GSN, Critchley JAJH. The estimation of paracetamol and its major metabolites in both plasma and urine by a single high-performance liquid chromatography assay. J Pharm Biomed Anal. 1994;12:1563–1572. doi: 10.1016/0731-7085(94)00859-0. [DOI] [PubMed] [Google Scholar]
- 20.Critchley JAJH, Nimmo GR, Gregson CA, Woolhouse NM, Prescott LF. Inter-subject and ethnic differences in paracetamol metabolism. Br J Clin Pharmacol. 1986;22:649–657. doi: 10.1111/j.1365-2125.1986.tb02953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burchell B, Coughtrie MWH. Genetic and environmental factors associated with variation of human xenobiotic glucuronidation and sulfation. Environ Health Perspect. 1997;105:739–747. doi: 10.1289/ehp.97105s4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 23.Smith EA, Macfarlane GT. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol. 1997;33:180–188. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- 24.Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2006;2:171–196. [Google Scholar]
- 25.Klaassen CD, Boles JW. The importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997;11:404–418. doi: 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- 26.Gamage N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 27.Morinaga Y, Fuke C, Arao T, Miyazaki T. Quantitative analysis of cresol and its metabolites in biological materials and distribution in rats after oral administration. Legal Med. 2004;6:32–40. doi: 10.1016/j.legalmed.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Schepers E, et al. p-Cresylsulphate, the main in vivo metabolite of p-cresol, activates leucocyte free radical production. Nephrol Dial Transplant. 2007;22:592–596. doi: 10.1093/ndt/gfl584. [DOI] [PubMed] [Google Scholar]
- 29.Martinez AW, Recht NS, Hostetter TH, Meyer TW. Removal of p-cresol sulphate by hemodialysis. J Am Soc Nephrol. 2005;16:3430–3436. doi: 10.1681/ASN.2005030310. [DOI] [PubMed] [Google Scholar]
- 30.de Loor H, Bammens B, Evenepoel P, De Preter V, Verbeke K. Gas-chromatographic-mass spectrometric analysis for measurement of p-cresol and its conjugated metabolites in uremic and normal serum. Clin Chem. 2005;51:1535–1538. doi: 10.1373/clinchem.2005.050781. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishna BS, et al. Estimation of phenolic conjugation by colonic mucosa. J Clin Pathol. 1989;42:620–623. doi: 10.1136/jcp.42.6.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34(Suppl 2):S12–S18. doi: 10.1016/s1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- 33.Galinsky RE, Corcoran GB. Influence of advanced age on the formation and elimination of acetaminophen metabolites by male rats. Pharmacology. 1986;32:313–320. doi: 10.1159/000138186. [DOI] [PubMed] [Google Scholar]
- 34.Rowland I. The influence of the gut microflora on food toxicity. Proc Nutr Soc. 1981;40:67–74. doi: 10.1079/pns19810011. [DOI] [PubMed] [Google Scholar]
- 35.Rowland IR. Reduction by the gut microflora of animals and man. Biochem Pharmacol. 1986;35:27–32. doi: 10.1016/0006-2952(86)90550-2. [DOI] [PubMed] [Google Scholar]
- 36.Rowland IR. Interactions of the gut microflora and the host in toxicology. Toxicol Pathol. 1988;16:147–153. doi: 10.1177/019262338801600207. [DOI] [PubMed] [Google Scholar]
- 37.Mikov M. The metabolism of drugs by the gut flora. Eur J Drug Metab Pharmacokinet. 1994;19:201–207. doi: 10.1007/BF03188922. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson JK, Wilson ID. Understanding ‘global’ systems biology: Metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 40.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 41.Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: A potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 42.Whitcomb DC, Block GD. Association of acetaminophen hepatotoxicity with fasting and ethanol use. J Am Med Assoc. 1994;272:1845–1850. doi: 10.1001/jama.1994.03520230055038. [DOI] [PubMed] [Google Scholar]
- 43.Kawakami K, Kojima K, Makino I, Kato I, Onoue M. Fasting enhances p-cresol production in the rat intestinal tract. Exp Anim. 2007;56:301–307. doi: 10.1538/expanim.56.301. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka E, Yamazaki K, Misawa S. Update: The clinical importance of acetaminophen hepatotoxicity in non-alcoholic and alcoholic subjects. J Clin Pharm Ther. 2000;25:325–332. doi: 10.1046/j.1365-2710.2000.00301.x. [DOI] [PubMed] [Google Scholar]
- 45.Thompson DC, Perera K, Fisher R, Brendel K. Cresol isomers: Comparison of toxic potency in rat liver slices. Toxicol Appl Pharmacol. 1994;125:51–58. doi: 10.1006/taap.1994.1048. [DOI] [PubMed] [Google Scholar]
- 46.Thompson DC, Perera K, London R. Quinone methide formation from para isomers of methylphenol (cresol), ethylphenol, and isopropylphenol: Relationship to toxicity. Chem Res Toxicol. 1995;8:55–60. doi: 10.1021/tx00043a007. [DOI] [PubMed] [Google Scholar]
- 47.Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 48.Coughtrie MWH. Sulfation through the looking glass—recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- 49.Kauffman FC. Sulfonation in pharmacology and toxicology. Drug Metab Rev. 2004;36:823–843. doi: 10.1081/dmr-200033496. [DOI] [PubMed] [Google Scholar]
- 50.Van Der Marel C, Anderson BJ, Tibboel D, Holford NHG. Modelling paracetamol urine metabolites. Paediatr Anaesth. 2002;12:827. [Google Scholar]
- 51.Hempel N, Gamage N, Martin JL, McManus ME. Human cytosolic sulfotransferase SULT1A1. Int J Biochem Cell Biol. 2007;39:685–689. doi: 10.1016/j.biocel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Ling WH, Hänninen O. Shifting from a conventional diet to an uncooked vegan diet reversibly alters fecal hydrolytic activities in humans. J Nutr. 1992;122:924–930. doi: 10.1093/jn/122.4.924. [DOI] [PubMed] [Google Scholar]
- 53.Sivsammye G, Sims HV. Presumptive identification of Clostridium difficile by detection of p-cresol in prepared peptone yeast glucose broth supplemented with p-hydroxyphenylacetic acid. J Clin Microbiol. 1990;28:1851–1853. doi: 10.1128/jcm.28.8.1851-1853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr. 1976;29:1448–1454. doi: 10.1093/ajcn/29.12.1448. [DOI] [PubMed] [Google Scholar]
- 55.Johnson GA, Barsuhn KJ, McCall JM. Sulfation of minoxidil by liver sulfotransferase. Biochem Pharmacol. 1982;31:2949–2954. doi: 10.1016/0006-2952(82)90268-4. [DOI] [PubMed] [Google Scholar]
- 56.Kim SY, et al. Formation of tamoxifen-DNA adducts via O-sulfonation, not O-acetylation, of α-hydroxytamoxifen in rat and human livers. Drug Metab Dispos. 2005;33:1673–1678. doi: 10.1124/dmd.105.005330. [DOI] [PubMed] [Google Scholar]
- 57.Thomas NL, Coughtrie MWH. Sulfation of apomorphine by human sulfotransferases: Evidence of a major role for the polymorphic phenol sulfotransferase, SULT1A1. Xenobiotica. 2003;33:1139–1148. doi: 10.1080/00498250310001609192. [DOI] [PubMed] [Google Scholar]
- 58.Adams RF, Murray KE, Earl JW. High levels of faecal p-cresol in a group of hyperactive children. Lancet. 1985;326:1313. doi: 10.1016/s0140-6736(85)91603-4. [DOI] [PubMed] [Google Scholar]
- 59.Cao L, Kirk MC, Coward LU, Jackson P, Whitaker JN. p-Cresol sulphate is the dominant component of urinary myelin basic protein like material. Arch Biochem Biophys. 2000;377:9–21. doi: 10.1006/abbi.2000.1764. [DOI] [PubMed] [Google Scholar]
- 60.Jackson PL, Cao L, Blalock JE, Whitaker JN. The requirement of ammonium or other cations linked with p-cresol sulfate for cross-reactivity with a peptide of myelin basic protein. Arch Biochem Biophys. 2003;418:119–124. doi: 10.1016/j.abb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Steventon GB, et al. Metabolism of low-dose paracetamol in patients with chronic neurological disease. Xenobiotica. 1990;20:117–122. doi: 10.3109/00498259009046818. [DOI] [PubMed] [Google Scholar]
- 62.Bradley H, Waring RH, Emery P, Arthur V. Metabolism of low-dose paracetamol in patients with rheumatoid arthritis. Xenobiotica. 1991;21:689–693. doi: 10.3109/00498259109039509. [DOI] [PubMed] [Google Scholar]
- 63.Alberti A, Pirrone P, Elia M, Waring RH, Romano C. Sulfation deficit in “low-functioning” autistic children: A pilot study. Biol Psychiatry. 1999;46:420–424. doi: 10.1016/s0006-3223(98)00337-0. [DOI] [PubMed] [Google Scholar]
- 64.Horvath K, Perman JA. Autism and gastrointestinal symptoms. Curr Gastroenterol Rep. 2002;4:251–258. doi: 10.1007/s11894-002-0071-6. [DOI] [PubMed] [Google Scholar]
- 65.Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parracho HMRT, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 67.Strang RR. The association of gastro-duodenal ulceration and Parkinson's disease. Med J Aust. 1965;1:842–843. doi: 10.5694/j.1326-5377.1965.tb72277.x. [DOI] [PubMed] [Google Scholar]
- 68.Goodhart PJ, DeWolf WE, Kruse LI. Mechanism-based inactivation of dopamine beta-hydroxylase by p-cresol and related alkylphenols. Biochem. 1987;26:2576–2583. doi: 10.1021/bi00383a025. [DOI] [PubMed] [Google Scholar]
- 69.Elliott AA, Elliot JR. Voltage-dependent inhibition of RCK1 K+ channels by phenol, p-cresol, and benzyl alcohol. Mol Pharmacol. 1997;51:475–483. [PubMed] [Google Scholar]
- 70.Vanholder R, De Smet R, Lesaffer G. p-Cresol: A toxin revealing many neglected but relevant aspects of uraemic toxicity. Nephrol Dial Transplant. 1999;14:2813–2815. doi: 10.1093/ndt/14.12.2813. [DOI] [PubMed] [Google Scholar]
- 71.Gaikwad NW, Bodell WJ. Formation of DNA adducts by microsomal and peroxidase activation of p-cresol: Role of quinone methide in DNA adduct formation. Chem Biol Interact. 2001;138:217–229. doi: 10.1016/s0009-2797(01)00274-5. [DOI] [PubMed] [Google Scholar]
- 72.Calderón-Guzmán D, et al. Effect of toluene and cresols on Na+,K+-ATPase, and serotonin in rat brain. Regul Toxicol Pharmacol. 2005;41:1–5. doi: 10.1016/j.yrtph.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Yan Z, et al. Bioactivation of 4-methylphenol (p-cresol) via cytochrome P450-mediated aromatic oxidation in human liver microsomes. Drug Metab Dispos. 2005;33:1867–1876. doi: 10.1124/dmd.105.006387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.