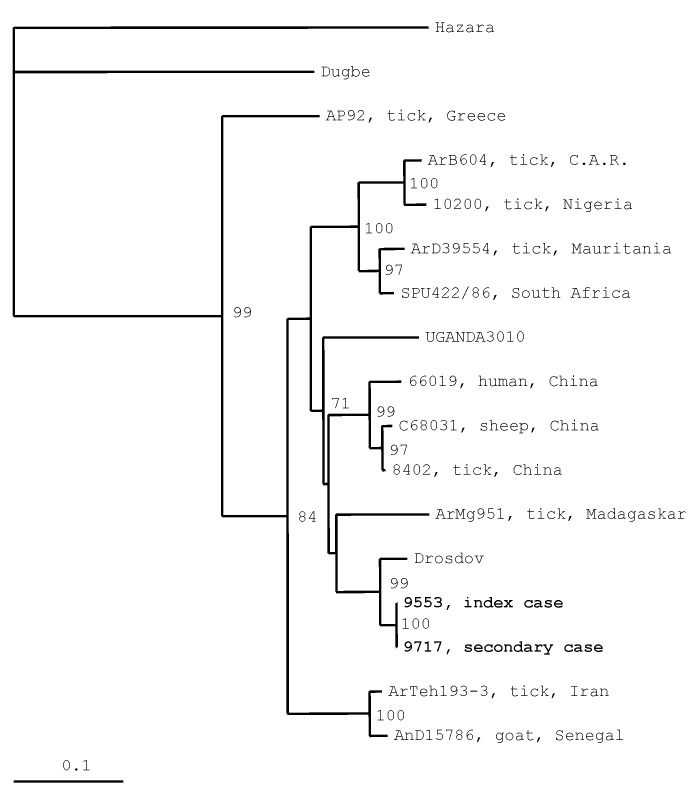
Figure.
Phylogenetic relationships based on 255-nt fragment from the small RNA segment between sequences obtained from this study and respective representative Crimean-Congo hemorrhagic fever strains from GenBank. In the phylogenetic tree, sequences of other two nairoviruses, Dugbe and Hazara, were included; Hazara virus was used as outgroup. The numbers indicate percentage bootstrap replicates (of 100) calculated by using SEQBOOT, DNADIST, FITCH, and CONSENSE from the PHYLIP package (7); values <70% are not shown. Horizontal distances are proportional to the nucleotide differences. The scale bar indicates 10% nucleotide sequence divergence. Vertical distances are for clarity only.