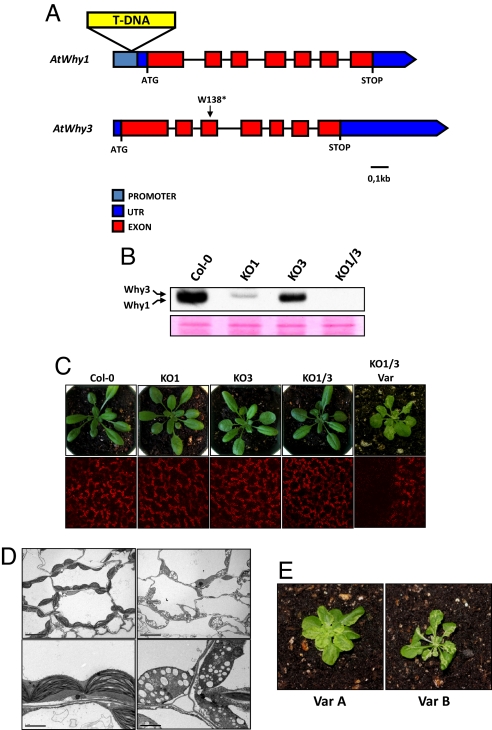
Fig. 1.
AtWhy1 and AtWhy3 are involved in the biogenesis of chloroplasts. (A) Physical maps of the AtWhy1 (AT1G14410) and AtWhy3 (AT2G02740) genes. The position of the T-DNA insertion in the KO1 line is indicated. KO3 is a TILLING line with a mutation that changes a TGG codon to a TGA stop codon in the AtWhy3 gene. An asterisk indicates the position of the mutation. (B) Protein gel blot analysis of simple and double ptWhirlies KO plants. Crude plastid proteins were separated by SDS/PAGE on a 15% polyacrylamide gel. Whirlies were detected by using an anti-AtWhy1/3 antibody. A section of the blot stained with Ponceau red is presented below as a loading control. (C) (Upper) Four-week-old individuals of the indicated genotypes are shown. (Lower) Fluorescence of chlorophyll was visualized by confocal microscopy. (D) Transmission electron microscopy of sections from green (Left) and white (Right) sectors of variegated leaves of KO1/3 plants. (Scale bars: 10 μm in Upper; 2 μm in Lower.) (E) Variegation phenotype varies between independent lines. Four-week-old individuals from the 2 variegated lines Var A (Left) and Var B (Right) are shown.