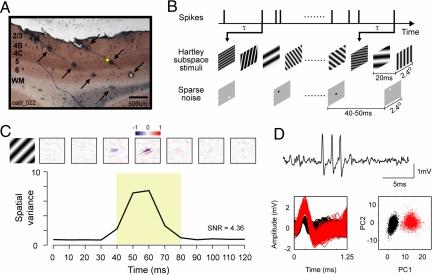
Fig. 1.
Mapping with Hartley stimuli and sparse noise and histological labeling of cell layers in V1. (A) A 50-μm cytochrome oxidase-stained brain section with multiple lesions (indicated by arrows) made along 2 recording tracks with quartz platinum/tungsten microelectrodes. (B) The spatiotemporal receptive fields were mapped with Hartley subspace stimuli and sparse noise and calculated by reverse correlation at different time delays (τ). (C) The spatial maps (Upper) and spatial variances (Lower) as a function of time for an example layer 4C simple cell (the yellow dot in A represents the place where the cell was recorded). The spatial maps were normalized by the response at the time when the spatial variance reached the peak. On subregions are shown in red (positive values), and off subregions are in blue (negative values). (D) A short segment of raw data (Upper) for the example layer 4C cell. For spike sorting, we first included waveforms with peaks >0.5 mV and then aligned them at their peaks at 0.25 ms to perform a principal components analysis. (Lower Left) Subsamples of waveforms from a 360-s segment (red: sorted spikes; black: noise). (Lower Right) A scatter plot of the dot product of each waveform with the first and second principal components (PC1 and PC2) of an average waveform from the 360-s segment.