Abstract
Static site-specific protein-DNA photocrosslinking permits identification of protein-DNA interactions within multiprotein-DNA complexes. Kinetic site-specific protein-DNA photocrosslinking--involving rapid-quench-flow mixing and pulsed-laser irradiation--permits elucidation of pathways and kinetics of formation of protein-DNA interactions within multiprotein-DNA complexes. We present detailed protocols for application of static and kinetic site-specific protein-DNA photocrosslinking to bacterial transcription initiation complexes.
Keywords: structure, kinetics, protein-DNA interaction, transcription initiation, RNA polymerase, promoter, open complex, phosphate, phosphorothioate, phenyl-azide photoactivatible crosslinking agent, quench-flow rapid mixing, pulsed-laser UV-irradiations
1. Introduction
1.1. Static site-specific protein-DNA photocrosslinking
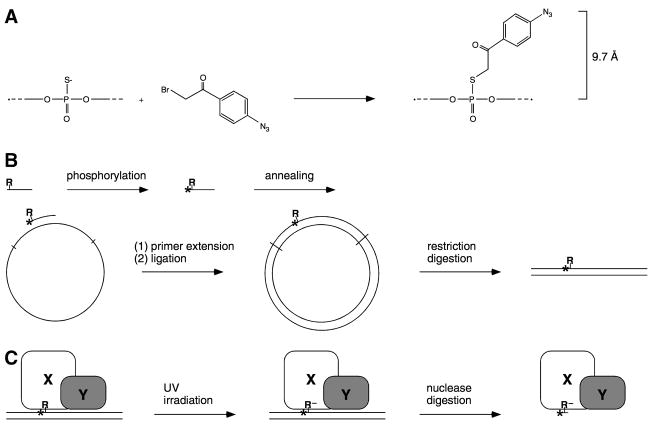
In published work, we have developed a procedure for site-specific protein-DNA photocrosslinking to define positions of proteins relative to DNA in protein-DNA and multiprotein-DNA complexes (1–6). The procedure has four parts (Figure 1):
Figure 1.
Site-specific protein-DNA photocrosslinking (1–6). (A,B) Chemical and enzymatic reactions are used to prepare a full-length-promoter DNA fragment with a phenyl-azide photoactivatible crosslinking agent (R) and an adjacent radioactive phosphorus (*) incorporated at a single, defined site. Based on the chemistry of incorporation, the maximum distance between the site of incorporation and the photoreactive atom is 9.7 Å; the maximum distance between the site of incorporation and a crosslinked atom is ~11 Å. (C) UV-irradiation of the derivatized protein-DNA complex initiates crosslinking. Nuclease digestion eliminates uncrosslinked DNA and converts crosslinked, radiolabelled DNA to a crosslinked, radiolabelled 3–5 nucleotide “tag.”
Chemical (7–9) and enzymatic (10) reactions are used to prepare a DNA fragment containing a photoactivatible crosslinking agent and an adjacent radiolabel incorporated at a single, defined DNA phosphate (with a 9.7 Å linker between the photoreactive atom of the crosslinking agent and the phosphorus atom of the phosphate, and with an ~11 Å maximum “reach” between potential crosslinking targets and the phosphorus atom of the phosphate).
The multiprotein-DNA complex of interest is formed using the site-specifically derivatized DNA fragment, and the multiprotein-DNA complex is UV-irradiated, initiating covalent crosslinking with proteins in direct physical proximity to the photoactivatible crosslinking agent.
Extensive nuclease digestion is performed, eliminating uncrosslinked DNA and converting crosslinked DNA to a crosslinked, radiolabelled 3–5 nucleotide “tag.”
The “tagged” proteins are identified.
The procedure is performed in systematic fashion, with preparation and analysis of at least 10 derivatized DNA fragments, each having the photoactivatible crosslinking agent incorporated at a single, defined DNA phosphate (typically each second DNA phosphate--each 12 Å--on each DNA strand spanning the region of interest; 1–6,11–15).
The results of the procedure define the translational positions of proteins relative to the DNA sequence. Plotted on a three-dimensional representation of a DNA helix, the results also define the rotational orientations of proteins relative to the DNA helix axis, and the groove orientations of proteins relative to the DNA major and minor grooves (1–5,11–13).
The procedure has been validated in experiments with three multiprotein-DNA complexes for which crystallographic structures are available: i.e., the TBP-DNA complex, the TBP-TFIIA-DNA complex, and the TBP-TFIIB-DNA complex (1,16–20). In each case, there was a one-for-one correspondence between sites at which strong crosslinking was observed and sites that in the crystallographic structure were within 11 Å of crosslinked proteins (1,16–20). The procedure also has been applied to multiprotein-DNA complexes for which crystallographic structures are not available (1–6,11–15), including bacterial transcription complexes containing 6–8 distinct polypeptides and having molecular masses of 450–400 kDa (5), archaeal transcription complexes containing 14 distinct polypeptides and having molecular masses of 400 kDa (12–14), and eukaryotic transcription complexes containing 16–27 distinct polypeptides and having molecular masses of 800–1700 kDa (2,4,15)
The procedure is related to a procedure developed by Geiduschek and co-workers (21–24; see also 25–30), but offers important advantages. First, since the photoactivatible crosslinking agent is incorporated into DNA chemically, it can be incorporated at a single, defined site. [In the procedure of Geiduschek and co-workers, this is true only at certain DNA sequences.] Second, since the photoactivatible crosslinking agent is incorporated on the DNA phosphate backbone, it can be incorporated at any nucleotide: A, T, G, or C. Third, since the photoactivatible crosslinking agent is incorporated on the DNA phosphate backbone, it probes interactions both in the DNA minor groove and in the DNA minor groove.
1.2. Kinetic site-specific protein-DNA photocrosslinking
The procedure for site-specific protein-DNA photocrosslinking summarized in section 1.1 can be combined with quench-flow rapid mixing and pulsed-laser flash UV-irradiation in order to permit analysis of kinetics of formation or breakage of protein-DNA interactions within protein-DNA and multiprotein-DNA complexes (“kinetic site-specific protein-DNA photocrosslinking”; S.D. and R.H.E, in preparation). Kinetic site-specific protein-DNA photocrosslinking involves four main steps (Figures 1,2):
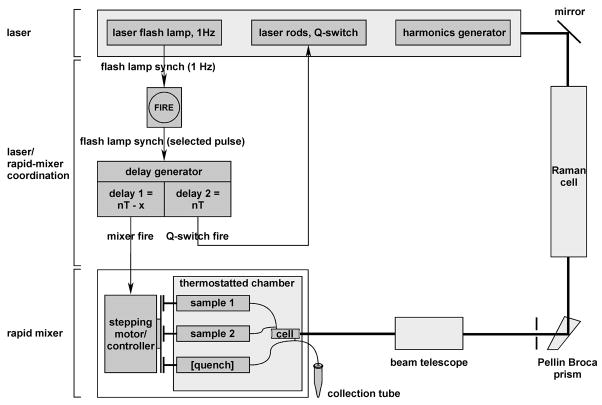
Figure 2.
Apparatus for kinetic site-specific protein-DNA photocrosslinking (S.D. and R.H.E., in preparation). The apparatus consists of a nanosecond pulsed Nd-YAG laser with integral third harmonic generator; a quench-flow rapid mixer; electronics for coordination of the laser and rapid mixer; and a Raman cell, Pellin-Broca prism, 355 nm laser optics (first mirror), and 309 nm laser optics (all other optics). To minimize variations in energy output, the laser flash lamp is allowed to operate continuously, and firing of the rapid mixer and firing of the laser Q-switch are coordinated with the timing of the laser flash lamp. Upon selection of a synch pulse from the laser flash lamp, the rapid mixer is fired at time = nT - x (where n is an integer, T is the flash lamp period, and x is the desired reaction time) resulting in mixing, the Q-switch is fired at time = nT resulting in UV-irradiation, and the rapid mixer is fired again at time = nT + y (where y is the desired post-UV-irradiation, pre-quenching reaction time; <1 ms in this work) resulting in quenching.
A DNA fragment containing a phenyl-azide photoactivatible crosslinking agent and an adjacent radiolabel incorporated at a single, defined DNA phosphate (prepared as summarized in section 1.1) is mixed with the protein(s) of interest at time = 0 using a quench-flow rapid mixer.
The sample is UV-irradiated at time = x using a single pulse from a pulsed laser, initiating covalent crosslinking with protein(s) in direct physical proximity to the phenyl-azide photoactivatible crosslinking agent.
The sample is mixed with a quench solution at time = x + ~1 ms, terminating covalent crosslinking (by inactivating photogenerated reactive species and by dissociating complexes).
Crosslinked protein(s) are identified (performed as summarized in section 1.1), and yield(s) of crosslinked protein(s) are quantified.
The procedure provides a “snapshot” or “motion-picture frame” of protein-DNA interactions at one position in DNA at one point in time. By performing a series of experiments with UV-irradiation at times x1, x2, …xn, one obtains a series of “motion-picture frames” and thus obtains a “cinematographic” record of protein-DNA interactions at one position in DNA. By performing such experiments systematically, using a set of DNA fragments derivatized at different single positions in DNA, one is able to define the full pathway and kinetics for formation or breakage of protein-DNA interactions within a protein-DNA or multiprotein-DNA complex.
1.3. Bacterial transcription initiation complexes
Escherichia coli RNA polymerase holoenzyme (RNAP) consists of two copies of an α subunit (37 kDa), one copy of a β subunit (150 kDa), one copy of a β′ subunit (160 kDa), one copy of an ω subunit (10 kDa), and one copy of a σ subunit (70 kDa for the principle σ subunit species, σ70) (31,32). RNAP is a molecular machine that carries out a complex series of reactions in transcription initiation (31,32). Formation of a catalytically competent transcription initiation complex involves at least three steps: (i) RNAP binds to promoter DNA to yield an RNAP-promoter closed complex; (ii) RNAP clamps tightly onto promoter DNA, to yield an RNAP-promoter intermediate complex; and (iii) RNAP unwinds ~14 bp of promoter DNA surrounding the transcription start, rendering accessible the genetic information in the template strand of DNA, to yield an RNAP-promoter open complex.
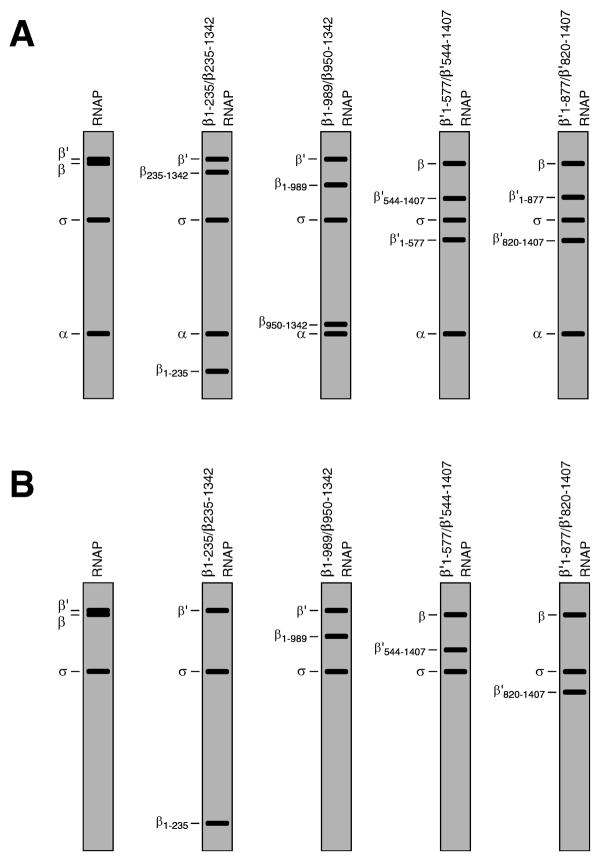
In published work, we have used static site-specific protein-DNA photocrosslinking to define the structural organization of the RNAP-promoter open complex (5,32). We constructed a set of 120 derivatized DNA fragments, each containing a photoactivatible crosslinking agent incorporated at a single, defined position of the lacPUV5 promoter (positions −95 to +25 relative to the transcription start site). For each derivatized DNA fragment, we formed the RNAP-promoter open complex, isolated the complex using non-denaturing polyacrylamide gel electrophoresis, UV-irradiated the complex in situ--in the gel matrix--and identified crosslinked polypeptides. We performed experiments both with wild-type RNAP and with RNAP derivatives having discontinuous β and β′ subunits (“split-β RNAP” and “split-β′ RNAP”; reconstituted in vitro from recombinant α, recombinant σ70, and sets of recombinant fragments of β and β′; see 33,34). Use of split-β and split-β′ RNAP permitted unambiguous assignment of crosslinks to β and β′ (which were not well resolved in SDS-polyacrylamide gel electrophoresis) and permitted rapid, immediate mapping of crosslinks to segments of β and β (e.g., N-terminal segment, central segment, or C-terminal segment) (Figure 3).
Figure 3.
Use of split-subunit RNAP derivatives permits unambiguous assignment of crosslinks to RNAP subunits and permits rapid mapping of crosslinks to segments of RNAP subunits (5; see 33,34). (A) Subunit compositions of RNAP, two split-β RNAP derivatives, and two split-β′ RNAP derivatives (idealized Coomassie-stained SDS-PAGE gels). (B) Results of site-specific protein-DNA photocrosslinking experiments using the RNAP derivatives of panel A and a DNA fragment derivatized at a site close to or in contact with residues 1–235 of β, residues 820–1407 of β′, and σ70 in the RNAP-promoter complex (idealized autoradiographs of SDS-PAGE gels).
In current work, we are using kinetic site-specific protein-DNA photocrosslinking to define the pathway and kinetics of formation of the RNAP-promoter open complex. We are analyzing a set of more than 30 derivatized DNA fragments, each containing a photoactivatible crosslinking agent incorporated at a single, defined position of the lacPUV5 promoter (with the sites being chosen to report key RNAP-promoter interactions spanning the RNAP-promoter interface).
In this chapter, we present protocols for use of static site-specific protein-DNA photocrosslinking to define the structural organization of the RNAP-promoter complex and for use of kinetic site-specific protein-DNA photocrosslinking to define the pathway and kinetics of formation of the RNAP-promoter open complex. In addition, we present support protocols for preparation of wild-type RNAP, split-β RNAP, and split-β′ RNAP.
2. Materials
2.1. Preparation of derivatized DNA fragment: chemical reactions
Azidophenacyl bromide (Sigma)
Tetraethylthiuram disulfide/acetonitrile (Applied Biosystems)
dA-CPG, dC-CPG, dG-CPG, T-CPG (1 μmol, 500 Å) (Applied Biosystems)
dA, dC, dG, T β-cyanoethylphosphoramidites (Applied Biosystems)
Reagent kit for oligodeoxyribonucleotide synthesis (0.02 M iodine) (Applied Biosystems)
Denaturing loading buffer (0.3% bromophenol blue, 0.3% xylene cyanol, 12 mM EDTA, in formamide)
0.5x TBE (45 mM Tris-borate, pH 8.3, 1 mM EDTA)
TE (10 mM Tris-HCl, pH 7.6, 1 mM EDTA)
50 mM triethylammonium acetate, pH 7.0 (Prime Synthesis)
1 M potassium phosphate, pH 7.0
3 M sodium acetate, pH 5.2
100% ethanol (store at −20°C)
70% ethanol (store at −20°C)
Dichloromethane (anhydrous) (Applied Biosystems)
Acetonitrile (anhydrous) (Applied Biosystems)
Acetonitrile (HPLC grade) (Fisher)
Formamide (Sigma)
12% polyacrylamide (29:1 acrylamide:bisacrylamide), 8 M urea, 0.5x TBE slab gel (10 × 7 × 0.075 cm)
Oligonucleotide purification cartridge (OPC) (Applied Biosystems)
LiChrospher 100 RP-18 reversed-phase HPLC column (5 μm) (Merck)
Autoradiography intensifying screen (Sigma)
254 nm germicidal lamp
ABI392 DNA/RNA synthesizer (Applied Biosystems)
Varian 5000 HPLC (Varian)
L-3000 diode-array HPLC UV detector (Hitachi)
Speedvac evaporator (Thermo Scientific)
2.2. Preparation of derivatized DNA fragment: enzymatic reactions
Derivatized oligodeoxyribonucleotide (Section 3.1)
M13mp2(ICAP-UV5) or M13mp2(ICAP-UV5)-rev ssDNA (see Notes 1,2)
T4 polynucleotide kinase (10 units/μl) (New England Biolabs, cat #M0201S)
T4 DNA polymerase (3 units/μl) (New England Biolabs, cat #M0203S)
T4 DNA ligase (5 units/μl) (Roche Applied Science, cat #799009)
HaeIII (40 units/μl)(Roche Applied Science, cat #1336029)
PvuII (40 units/μl)(Roche Applied Science, cat #899216)
[γ32P]-ATP (10 mCi/ml, 6000 Ci/mmol) (Perkin Elmer)
100 mM ATP (GE Life Sciences)
100 mM dNTPs (GE Life Sciences)
Downstream primer (5′-CGGTGCGGGCCTCTTCGCTATTAC-3′)
10x phosphorylation buffer (500 mM Tris-HCl, pH 7.6, 100 mM MgCl2, 15 mM β-mercaptoethanol)
10x annealing buffer (400 mM Tris-HCl, pH 7.9, 500 mM NaCl, 100 mM MgCl2)
10x digestion buffer (100 mM Tris-HCl, pH 7.9, 500 mM NaCl, 100 mM MgCl2) (see Note 3)
Elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, pH 7.5, 1 mM EDTA)
Denaturing loading buffer (0.3% bromophenol blue, 0.3% xylene cyanol, 12 mM EDTA, in formamide)
Nondenaturing loading buffer (0.3% bromophenol blue, 0.3% xylene cyanol, 30% glycerol, in water)
0.5x TBE (45 mM Tris-borate, pH 8.3, 1 mM EDTA)
TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA)
Low-EDTA TE (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA)
0.5 M EDTA, pH 8.0
10% SDS
100% ethanol (store at −20 °C)
70% ethanol (store at −20 °C)
12% polyacrylamide (29:1 acrylamide:bisacrylamide), 8 M urea, 0.5x TBE slab gel (10 × 7 × 0.075 cm)
7.5% polyacrylamide (29:1 acrylamide:bisacrylamide), 0.5x TBE slab gel (10 × 7 × 0.15 cm)
CHROMA SPIN+TE-10 spin column (Clontech)
CHROMA SPIN+TE-100 spin column (Clontech)
Spin-X centrifuge filter (0.22 μm, cellulose acetate) (Fisher)
PicoGreen dsDNA quantitation kit (Invitrogen, cat #P7589)
Disposable scalpels (VWR)
Autoradiography markers (Stratagene)
Light box (VWR)
Speedvac evaporator (Savant)
2.3. Preparation of RNAP and RNAP derivatives
E. coli strain XL1-blue (Stratagene, cat #200249)
E. coli strain BL21(DE3) pLysS (Novagen, cat #69388-3)
Plasmids encoding RNAP subunits (see Table 1)
Plasmids encoding fragments of RNAP subunits (see Table 2)
LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl; autoclave-sterilized)
TYE agar containing 200 μg/ml ampicillin and 35 μg/ml chloramphenicol (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotics after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 ml/plate)
TYE agar containing 200 μg/ml ampicillin and 20 μg/ml tetracycline (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotics after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 ml/plate)
TYE agar containing 40 μg/ml kanamycin and 35 μg/ml chloramphenicol (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotics after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 ml/plate)
TYE agar containing 40 μg/ml kanamycin and 20 μg/ml tetracycline (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotics after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 ml/plate)
100 mg/ml ampicillin (filter-sterilized) (Sigma)
35 mg/ml chloramphenicol in ethanol (filter-sterilized) (Sigma)
40 mg/ml kanamycin (filter-sterilized) (Sigma)
20 mg/ml tetracycline in methanol (filter-sterilized) (Sigma)
1 M IPTG (filter-sterilized) (Roche Applied Science)
Buffer A (20 mM Tris-HCl pH 7.9, 500 mM NaCl, 5 mM imidazole)
Buffer B (20 mM Tris-HCl pH 7.9, 6 M guanidine chloride, 500 mM NaCl)
Buffer C (40 mM Tris-HCl, pH 7.9, 300 mM KCl, 10 mM EDTA, 1 mM PMSF, 1 mM DTT)
Buffer D (50 mM Tris-HCl pH 7.9, 6 M guanidine chloride, 10 mM MgCl2, 0.01 mM ZnCl2, 1 mM EDTA, 10 mM DTT, 10% glycerol)
Buffer E (50 mM Tris-HCl pH 7.9, 200 mM KCl, 10 mM MgCl2, 0.01 mM ZnCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, 20% glycerol)
Buffer F (50 mM Tris-HCl pH 7.9, 5% glycerol)
α storage buffer (50 mM Tris-HCl, pH 7.9, 200 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, 20% glycerol)
2x SDS loading buffer (63 mM Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 25% glycerol, 0.3% bromophenol blue)
SDS running buffer (25 mM Tris, 250 mM glycine, pH 8.3, 0.1% SDS)
Destaining solution (10% acetic acid, 50% methanol, 40% water)
100 mM phenylmethanesulfonyl fluoride (PMSF) in ethanol (Sigma)
2% lysozyme (Sigma, cat #L-6876) (~50,000 units/mg)
10% sodium desoxycholate (Sigma)
10% n-octyl-β-D-glucopyranoside (Sigma)
Triton X-100 (Sigma)
2 M imidazole (pH adjusted to 8.0 with 10 M HCl) (Sigma)
Glycerol (Fisher)
Trichloroacetic acid (Aldrich)
Coomassie Brilliant Blue G-250 (Bio-Rad)
Acetone (Aldrich)
10% polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm)
Prestained protein molecular weight markers (7–210 kDa) (Bio-Rad)
Protein Assay Kit (Bio-Rad, cat #500–0002)
Ni:NTA-agarose (Qiagen)
Dialysis membranes (10 kDa molecular-weight cutoff) (VWR, cat #25223-821)
Dialysis-membrane closures (VWR)
Collodion dialysis bags (10 kDa molecular-weight cutoff) (Schleicher & Schuell)
Nanosep-30K centrifugal concentrators (VWR)
Econo-Pac 20 ml chromatography columns (Bio-Rad)
Chromatography column frits (1.5 × 0.3 cm) (Bio-Rad)
15 ml culture tubes (autoclave-sterilized) (VWR)
Culture-tube stainless steel closures (autoclave-sterilized) (VWR)
2.8 L triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass, Inc., cat #2551-02800)
30 ml polypropylene copolymer centrifuge tube with cap (VWR, cat #21010-567)
250 ml polypropylene copolymer centrifuge bottle with cap (VWR, cat #21020-028)
1 L polypropylene copolymer centrifuge bottle with cap (VWR, cat #21020-061)
200 ml steel beaker (VWR)
Branson 450 sonicator (VWR)
Sorvall RC-3B centrifuge (Thermo Scientific)
Sorvall RC-5B centrifuge (Thermo Scientific)
Table 1.
Plasmids encoding RNAP subunits
| Plasmid | Relevant Characteristics | Source |
|---|---|---|
| pHTT7f1-NHα | ApR; ori-pBR322; ori-f1; ϕ10P-rpoA(H6,Nter)a | 35,36 |
| pMKSe2 | ApR; ori-pBR322; lacP-rpoB | 37 |
| pT7β′ | ApR; ori-pBR322; ϕ10P-rpoC | 38 |
| pHTT7f1-α | ApR; ori-pBR322; ori-f1; ϕ10P-rpoD | 35,36 |
| pT7ω | ApR; ori-pBR322; ori-f1; ϕ10P-rpoZ | 39 |
rpoA(H6,Nter) is a derivative of rpoA having a non-native hexahistidine coding sequence immediately after the rpoA start codon.
Table 2.
Plasmids encoding fragments of RNAP subunits
| Plasmid | Relevant Characteristics | Source |
|---|---|---|
| pβ1–235 | ApR KmR; ori-pBR322; lacP-rpoB(1–235) | 33 |
| pβ235–1342 | ApR; ori-pBR322; lacP-rpoB(235–1342) | 33 |
| pβ1–989 | ApR; ori-pBR322; lacP-rpoB(1–989) | 33 |
| pβ950–1342 | ApR; ori-pBR322; ϕ10P-rpoB(950–1342) | 33 |
| pβ′1–577 | ApR; ori-pBR322; ori-f1; lacP-ϕ10P-rpoC(1–577) | 34 |
| pβ′544–1407 | ApR; ori-pBR322; ori-f1; lacP-ϕ10P-rpoC(544–1407) | 34 |
| pβ′1–877 | ApR; ori-pBR322; ori-f1; lacP-ϕ10P-rpoC(1–877) | 34 |
| pβ′820–1407 | KmR; ori-pBR322; ori-f1; ϕ10P-rpoC(820–1407) | 34 |
2.4. Static photocrosslinking
Cystamine dihydrochloride (Sigma)
Acryloyl chloride (Aldrich)
Acrylamide (Bio-Rad)
TEMED (BioRad)
10% ammonium persulfate (freshly made)
SurfaSil siliconizing agent (Pierce)
Derivatized promoter DNA fragment (Section 3.2)
RNAP or RNAP derivative (Section 3.3)
DNase I (126 units/μl) (Sigma, cat #D7291)
Micrococcal nuclease in nuclease dilution solution (50 units/μl) (GE Life Sciences, cat #27-0584)
Nuclease dilution solution (5 mM CaCl2, 0.1 mM PMSF, 50% glycerol)
2x DTT-free transcription buffer (50 mM HEPES-HCl, pH 8.0, 200 mM KCl, 20 mM MgCl2, 10% glycerol)
Nondenaturing loading buffer (0.3% bromophenol blue, 0.3% xylene cyanol, 30% glycerol)
5x SDS loading buffer (300 mM Tris-HCl, pH 6.8, 10% SDS, 20 mM EDTA, 25% β-mercaptoethanol, 0.1% bromophenol blue, 50% glycerol)
0.5x TBE (45 mM Tris-borate, pH 8.0, 1 mM EDTA)
SDS running buffer (25 mM Tris, 250 mM glycine, pH 8.3, 0.1% SDS)
10% SDS
1 M DTT (freshly made)
0.1 mM PMSF (Sigma)
0.22 μg/ml heparin (Sigma, cat #H-3393) (grade I-A, from porcine intestinal mucosa, ~170 USP units/mg)
7–15% gradient polyacrylamide (37.5:1 acrylamide:bisacrylamide) Tris-HCl slab gel (Bio-Rad, cat #161-0902)
Prestained protein molecular weight markers (7–210 kDa) (Bio-Rad)
Silicone rubber heating mat (200 W, 120 VAC; 25 × 10 cm) (Cole-Parmer, cat #P-03125-40)
Variable voltage controller (Cole-Parmer, cat #P-01575-10)
Digital thermometer (Cole-Parmer, cat #P-91000-00)
Thermocouple probe (needle, 0.7 mm diameter) (Cole-Parmer, cat #P-08505-92)
Large binder clips (width = 5 cm) (Staples)
Filter unit (22 μm pore size, 250 ml) (Millipore)
50 ml Buchner funnel with glass frit (10 μm pore size) (Fisher)
500 ml separating funnel (Fisher)
X-ray exposure holder with intensifying screen (Kodak)
Light box (VWR)
Rayonet RPR-100 photochemical reactor equipped with 16 RPR-3500 Å tubes (Southern New England Ultraviolet)
Speedvac evaporator (Savant)
2.5. Kinetic photocrosslinking
1. Derivatized promoter DNA fragment (Section 3.2).
2. RNAP or RNAP derivative (Section 3.3).
3. DNase I (126 units/μl) (Sigma).
4. Micrococcal nuclease (2000 units/μl) (New England Biolabs).
5. Nuclease dilution solution: 5 mM CaCl2, 0.1 mM PMSF, and 50% glycerol.
6. OmniCleave endonuclease (200 units/μl) (Epicentre).
7. OmniCleave endonuclease dilution buffer (Epicentre).
8. DTT- and glycerol-free transcription buffer: 25 mM HEPES-HCl, pH 8.0, 100 mM KCl, 10 mM MgCl2, and 0.1% Tween-20.
9. DTT- and glycerol-free, BSA-containing transcription buffer: 25 mM HEPES-HCl, pH 8.0, 100 mM KCl, 10 mM MgCl2, 0.1% Tween-20, 100 μg/ml BSA (prepared immediately before use; see section 3.5.1).
10. Urea/NaI solution: 5 M urea and 0.5 M NaI
11. Quench solution: 5 M urea, 0.5 M NaI, 0.5 M β-mercaptoethanol (prepared immediately before use; see section 3.5.1).
12. 0.1 M PMSF in ethanol (Sigma).
13. Criterion precast 4–20% gradient Tris-HCl gel (Bio-Rad).
14. Precision-Plus protein dual-color standards (10–250 kDa) (Bio-Rad).
15. 5X SDS loading buffer: 300 mM Tris-HCl, pH 8.3, 10% SDS, 20 mM EDTA, 25% β-mercaptoethanol, 0.1% bromophenol blue, and 50% glycerol.
16. SDS running buffer: 25 mM Tris, 250 mM glycine, pH 8.3, and 0.1% SDS.
17. Tween-20 (Bio-Rad).
18. Bovine serum albumin (BSA; Sigma).
19. Urea (Fisher).
20. NaI (Fisher).
21. β-mercaptoethanol (14.2 M; Bio-Rad).
22. Methanol (Fisher).
23. Argon (compressed, high-purity; Airgas).
24. 50 ml polypropylene centrifuge tubes (Falcon, Fisher).
25. 5 ml Luer-Lok disposable syringes (VWR).
26. 1 ml tuberculin disposable syringes (VWR).
27. Hamilton 50 μl Luer-Lok syringe (Hamilton).
28. Linagraph direct print paper (Kodak).
29. Microprocessor-controlled, stepping-motor-driven rapid-quench-flow system equipped with 20 μl sample chamber with acrylic OP1 optical windows(RQF-3 with modifications; KinTek, Austin TX) (see Note 21).
30. Nanosecond pulsed Nd-YAG laser equipped with third-harmonic generator (Powerlite 7010-DS/TS/QS; 1 Hz; Continuum, Santa Clara CA) (see Note 22).
31. Raman cell (RC-1; Light Age, Somerset NJ).
32. Pellin-Broca prism (CVI Laser, Albuquerque NM).
33. 355 nm and 309 nm laser optics (CVI Laser, Albuquerque NM).
34. Laser optics mounts (Thorlabs, Newton NJ).
34. Optical table (4′ × 6′; Newport, Irvine CA).
35. Laser calorimeter (AC25UV; Scientech, Boulder CO).
36. Laser energy meter (Astral AD30; Scientech, Boulder CO).
37. Laser safety goggles (OD ≥6 at 1064 nm; OD ≥7 at 190–532 nm; L332CB; Uvex Safety, Inc., Smithfield RI).
38. Digital delay/pulse generator (DG535; Stanford Research Systems, Sunnyvale CA).
39. Circulating water bath (Isotemp 1016D; Fisher).
40. Multimode imager (Typhoon 9400; GE Life Sciences).
3. Methods
3.1. Preparation of derivatized DNA fragment: chemical reactions
3.1.1. Preparation of phosphorothioate oligodeoxyribonucleotide
Perform 24 standard cycles of solid-phase β-cyanoethylphosphoramidite oligodeoxyribonucleotide synthesis to prepare CPG-linked precursor containing residues 3–26 of desired oligodeoxyribonucleotide. Use the following settings: cycle, 1.0 μmol CE; DMT, on; end procedure, manual.
Replace iodine/water/pyridine/tetrahydrofuran solution (bottle 15) by tetraethylthiuram disulfide/acetonitrile solution. Perform one modified cycle of solid-phase β-cyanoethylphosphoramidite oligodeoxyribonucleotide synthesis to add residue 2 and phosphorothioate linkage. Use the following settings: cycle, 1.0 μmol sulfur; DMT, on; end procedure, manual.
Replace tetraethylthiuram disulfide/acetonitrile solution (bottle 15) by iodine/water/pyridine/tetrahydrofuran solution. Place collecting vial on the DNA synthesizer. Perform one standard cycle of solid-phase β-cyanoethylphosphoramidite oligodeoxyribonucleotide synthesis to add residue 1. Use the following settings: cycle, 1.0 μM CE; DMT, on; end procedure, CE.
Remove collecting vial, screw cap tightly, and de-block by incubating 8 hours at 55°C. Transfer sample to 6 ml polypropylene round-bottom tube, place tube in Speedvac and spin 20 min with Speedvac lid ajar and with no vacuum (allowing evaporation of ammonia). Close Speedvac lid, apply vacuum, and dry.
De-tritylate and purify ~75 nmol on OPC according to supplier’s protocol.
Dry in Speedvac.
Re-suspend in 100 μl TE. Remove 2 μl aliquot, dilute with 748 μl TE, and determine concentration from UV-absorbance at 260 nm (molar extinction coefficient = 240,000 AU M−1 cm−1).
To confirm purity of oligodeoxyribonucleotide, mix aliquot containing 1 nmol oligodeoxyribonucleotide with equal volume of formamide. Apply to 12% polyacrylamide (29:1 acrylamide:bisacrylamide), 8 M urea, 0.5x TBE slab gel (10 × 7 × 0.075 cm). As marker, load in adjacent lane 5 μl denaturing loading buffer. Electrophorese 30 min at 25 V/cm. Disassemble gel, place on intensifying screen, and view in dark using 254 nm germicidal lamp. Oligodeoxyribonucleotide should appear as dark shadow against green background and should migrate more slowly than bromophenol blue. If purity is 95%, proceed to next step.
Divide remainder of sample into 50 nmol aliquots, transfer to 1.5 ml siliconized polypropylene microcentrifuge tubes, dry in Speedvac, and store at −20°C (stable for at least 2 y).
3.1.2. Derivatization of oligodeoxyribonucleotide [all steps carried out under subdued lighting (see Note 4)]
Dissolve 10 mg (42 μmol) azidophenacyl bromide in 1 ml chloroform. Transfer 100 μl aliquots (4.2μmol) to 1.5 ml siliconized polypropylene microcentrifuge tubes, and dry in Speedvac. Wrap tubes with aluminum foil, and store desiccated at 4°C (stable indefinitely).
Resuspend 50 nmol aliquot of phosphorothioate oligodeoxyribonucleotide (Section 3.1.1) in 50 μl water, and re-suspend 42 μmol aliquot of azidophenacyl bromide in 220 μl methanol.
Mix 50 μl (50 nmol) phosphorothioate oligodeoxyribonucleotide solution, 5 μl 1 M potassium phosphate (pH 7.0), and 55 μl (1 μmol) azidophenacyl bromide solution in 1.5 ml siliconized polypropylene microcentrifuge tube. Incubate 3 h at 37°C in the dark.
Precipitate derivatized oligodeoxyribonucleotide by adding 11 μl 3M sodium acetate (pH 5.2), and 275 μl ice-cold 100% ethanol. Invert tube several times, and place at −80°C for 30 min. Centrifuge 5 min at 13,000 × g at 4°C. Remove supernatant, wash pellet with ice-cold 70% ethanol. Air dry 15 min at RT. Store at −20 °C (stable for at least 1 y).
3.1.3. Purification of derivatized oligodeoxyribonucleotide [all steps carried out under subdued lighting (see Note 4)]
Resuspend derivatized oligodeoxyribonucleotide in 100 μl 50 mM triethylammonium acetate (pH 7.0).
Analyze 5 μl aliquot by C18 reversed-phase HPLC to confirm efficiency of derivatization reaction. Use LiChrospher 100 RP-18 C18 reversed-phase HPLC column (5 μm), with solvent A = 50 mM triethylammonium acetate (pH 7.0), 5% acetonitrile; solvent B = 100% acetonitrile; and flow rate = 1 ml/min. Equilibrate column with 10 column volumes solvent A before loading sample. After loading sample, wash column with 6 column volumes solvent A, and elute with 50 min gradient of 0–70% solvent B in solvent A. Derivatized and underivatized oligodeoxyribonucleotides elute at ~25% solvent B and ~16% solvent B, respectively (see Notes 5,6).
If derivatization efficiency is 80%, purify remainder of sample using procedure of Step 2, collecting peak fractions (see Notes 5,6).
Pool peak fractions, divide into 1 ml aliquots, and dry in Speedvac. Store desiccated at −20°C in the dark (stable for at least 1 y).
Resuspend one aliquot in 100 μl TE. Remove 5 μl, dilute with 495 μl water, and determine concentration from UV-absorbance at 260 nm (molar extinction coefficient = 242,000 AU M−1 cm−1).
Divide remainder of derivatized-oligodeoxyribonucleotide/TE solution from Step 5 into twenty 5 pmol aliquots and one larger aliquot, dry in Speedvac, and store desiccated at −20°C in the dark (stable for at least 1 y).
3.2. Preparation of derivatized DNA fragment: enzymatic reactions
3.2.1. Radiophosphorylation of derivatized oligodeoxyribonucleotide [all steps carried out under subdued lighting (see Note 4)]
Re-suspend 5 pmol derivatized oligodeoxyribonucleotide in 12 μl water. Add 2 μl 10x phosphorylation buffer, 5 μl [γ32P]ATP (50 μCi) and 1 μl (10 units) T4 polynucleotide kinase. Incubate 15 min at 37°C. Terminate reaction by heating 5 min at 65°C (see Note 7).
Add 15 μl water.
Desalt radiophosphorylated derivatized oligodeoxyribonucleotide into TE using CHROMA SPIN+TE-10 spin column according to supplier’s protocol.
Immediately proceed to next step, or, if necessary, store radiophosphorylated derivatized oligodeoxyribonucleotide solution at −20°C in the dark (stable for up to 24 h).
3.2.2. Annealing, extension, and ligation of radiophosphorylated derivatized oligodeoxyribonucleotide [all steps carried out under subdued lighting (see Note 4)]
In 1.5 ml siliconized polypropylene microcentrifuge tube, mix 34 μl radiophosphorylated derivatized oligodeoxyribonucleotide, 1 μl 10 μM downstream primer, 1 μl 1 μM M13mp2(ICAP-UV5) ssDNA (for analysis of crosslinks to template DNA strand) or M13mp2(ICAP-UV5)-rev ssDNA (for analysis of crosslinks to non-template DNA strand), and 4 μl 10x annealing buffer.
Heat 5 min at 65°C (see Note 7). Transfer to 500 ml beaker containing 200 ml water at 65°C, and place beaker at room temperature to permit slow cooling (65°C to 25°C in ~60 min).
Add 1 μl 25 mM dNTPs, 1 μl 100 mM ATP, 1 μl (3 units) T4 DNA polymerase, and 1 μl (5 units) T4 DNA ligase. Perform parallel reaction without ligase as “no-ligase” control.
Incubate 15 min at room temperature, followed by 35 min at 37°C. Terminate reaction by adding 1 μl 10% SDS.
Desalt into TE using CHROMA SPIN+TE-100 spin column according to supplier’s protocol. Immediately proceed to next step.
3.2.3. Digestion and purification of derivatized DNA fragment [all steps carried out under subdued lighting (see Note 4)]
In 1.5 ml siliconized polypropylene microcentrifuge tube, mix 40 μl product from Section 3.2.2, 4.5μl 10x digestion buffer, 0.25 μl (10 units) HaeIII or 0.25 μl (10 units) PvuII (see Note 8). Incubate 1 h at 37°C.
Perform parallel reaction using 40 μl “no-ligase” control from Section 3.2.2.
Mix 3 μl aliquots of reaction of Step 1 and of “no-ligase” control reaction of Step 2, each with 7 μl denaturing loading buffer. Heat 5 min at 65°C, and then apply to 12% polyacrylamide (29:1 acrylamide:bisacrylamide), 8 M urea, 0.5x TBE slab gel (10 × 7 × 0.075 cm). As marker, load in adjacent lane 5 μl denaturing loading buffer. Electrophorese 30 min at 25 V/cm. Dry gel, expose to x-ray film 1 h at room temperature, and process film. Estimate ligation efficiency by comparing reaction and “no-ligase” control lanes. If ligation efficiency is 80%, proceed to next step. If not, repeat Steps 3.2.1 and 3.2.2.
Mix remainder of reaction of Step 1 (42 μl) with 10 μl 50% glycerol. Apply to nondenaturing 7.5% polyacrylamide (29:1 acrylamide:bisacrylamide), 0.5x TBE slab gel (10 × 7 × 0.15 cm). As marker, load in adjacent lane 5 μl nondenaturing loading buffer. Electrophorese at 25 V/cm until bromophenol blue reaches bottom of the gel.
Remove one glass plate, and cover gel with plastic wrap. Attach two autoradiography markers to gel. Expose to x-ray film for 60–90 s at room temperature, and process film. Cut out portion of film corresponding to derivatized DNA fragment. Using light box, superimpose cut-out film on gel, using autoradiography markers as alignment reference points. Using disposable scalpel, excise portion of gel corresponding to derivatized DNA fragment.
Place excised gel slice in 1.5 ml siliconized polypropylene microcentrifuge tube, and crush with 1 ml pipette tip. Add 300 μl elution buffer, centrifuge 5 s at 5,000 × g, and incubate 12 h at 37°C.
Transfer supernatant to Spin-X centrifuge filter, centrifuge 1 min at 13,000 × g at room temperature in fixed-angle microcentrifuge.
Transfer filtrate to 1.5 ml siliconized polypropylene microcentrifuge tube. Precipitate derivatized DNA fragment by addition of 1 ml ice-cold 100% ethanol. Invert tube several times, and place at −20°C for 30 min. Centrifuge 5 min at 13,000 × g at 4°C in fixed-angle microcentrifuge. Remove and dispose of supernatant, wash pellet with 500 μl ice-cold 70% ethanol, and air dry 15 min at room temperature.
Re-suspend in 30 μl low-EDTA TE. Determine radioactivity by Cerenkov counting. Remove 1 μl aliquot, and determine DNA concentration using PicoGreen dsDNA quantitation kit according to supplier’s protocol. Calculate specific activity (expected specific activity ~5,000 Ci/mmol).
Store derivatized DNA fragment at 4°C in the dark (stable for ~1 week).
3.3. Preparation of RNAP and RNAP derivatives
3.3.1. Preparation of hexahistidine-tagged recombinant α subunit
Transform E. coli strain BL21(DE3) pLysS with plasmid pHTT7f1-NHα. Plate to TYE agar containing 200 μg/ml ampicillin and 35 μg/ml chloramphenicol, and incubate 12 h at 37°C.
Inoculate single colony into 5 ml LB containing 200 μg/ml ampicillin and 35 μg/ml chloramphenicol in 15 ml culture tube with culture tube stainless steel closure, and shake vigorously for 12 h at 37°C. Transfer to 15 ml polypropylene centrifuge tube, and centrifuge 5 min at 3,000 × g at room temperature. Discard supernatant, wash cell pellet twice with 5 ml LB, and re-suspend cell pellet in 5 ml LB in 15 ml polypropylene centrifuge tube.
Inoculate into 1 L LB containing 200 μg/ml ampicillin and 35 μg/ml chloramphenicol in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 1 ml 1 M IPTG, and shake vigorously for an additional 3 h at 37°C.
Transfer culture to 1 L polypropylene copolymer centrifuge bottle. Harvest cells by centrifugation 20 min at 5,000 × g at 4°C.
Resuspend cell pellet in 100 ml buffer A at 4°C. Transfer into 200 ml steel beaker, and place beaker on ice. Lyse cells with four 40 s sonication pulses at 25% maximum sonicator output (2 min pause between each pulse).
Transfer lysate to 250 ml polypropylene copolymer centrifuge bottle. Centrifuge 15 min at 15,000 × g at 4°C. Collect supernatant.
Transfer supernatant to 250 ml glass beaker. Add 35 g ammonium sulfate, and stir 20 min on ice.
Transfer suspension to 250 ml polypropylene copolymer centrifuge bottle. Centrifuge 20 min at 15,000 × g at 4°C. Discard supernatant.
Resuspend pellet in 28 ml buffer B containing 5 mM imidazole. Transfer to 30 ml polypropylene copolymer centrifuge tube, and rock gently 30 min at 4°C. Centrifuge 15 min at 15,000 × g at 4°C.
Load supernatant onto 5 ml Ni:NTA-agarose column pre-equilibrated with 25 ml buffer B containing 5 mM imidazole (prepared by pouring 10 ml Ni:NTA-agarose suspension into a 20 ml Econo-Pac column, removing snap-off tip at bottom of column, allowing liquid to drain, and then placing a frit on the top of column bed). Collect flow-through, and reload onto column. Wash column with 50 ml buffer B containing 5 mM imidazole, and 25 ml buffer B containing 10 mM imidazole. Elute column with 15 ml buffer B containing 20 mM imidazole, 15 ml buffer B containing 30 mM imidazole, 15 ml buffer B containing 40 mM imidazole, and 15 ml buffer B containing 150 mM imidazole. Collect 5 ml fractions.
Transfer 10 μl aliquot of each fraction to 1.5 ml siliconized polypropylene microcentrifuge tube, add 90 μl water and 100 μl 10% trichloroacetic acid. Place on ice 20 min. Centrifuge 5 min at 13,000 × g at room temperature. Discard supernatant. Wash pellet with 500 μl acetone, and air dry for 15 min. Dissolve pellet in 5 μl water, add 5 μl 2x SDS loading buffer, heat 3 min at 100°C, and apply to 10% polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm). As marker, load into adjacent lane 5 μl prestained protein molecular weight markers. Electrophorese in SDS running buffer at 25 V/cm until bromophenol blue reaches bottom of gel. Stain gel by gently shaking for 5 min in 50 ml 0.2% Coomassie Brilliant Blue G-250 in destaining solution. Destain by gently shaking for 1 h in 100 ml destaining solution.
Pool fractions containing homogenous α (typically fractions with buffer B containing 40–150 mM imidazole). Dialyze using 10 kDa molecular-weight-cutoff dialysis membrane against two 1 L changes of α storage buffer for 16 h at 4°C.
Determine protein concentration and total protein amount using Bio-Rad Protein Assay according to supplier’s protocol.
After dialysis, measure volume, and transfer to 30 ml polypropylene copolymer centrifuge tube. Add 3 g ammonium sulfate per 10 ml, and rock gently 20 min at 4°C. Centrifuge 20 min at 15,000 × g at 4°C.
Remove and discard 10 ml of supernatant. Resuspend pellet in remaining supernatant. Divide into 50 μl aliquots, and transfer to 1.5 ml siliconized polypropylene microcentrifuge tubes. Centrifuge aliquots 5 min at 13,000 × g at 4°C. Store at −80°C (stable for at least 1 y). Expected yield: 20–30 mg (250–500 μg/aliquot). Expected purity: >99%.
3.3.2. Preparation of crude recombinant RNAP subunits and subunit fragments
Transform plasmid encoding RNAP subunit or subunit fragment into E. coli strain BL21(DE3) pLysS (for plasmids with ϕ10P- or lacP-ϕ10P-based expression; Tables 1, 2) or E. coli strain XL1-blue (for plasmids with lacP-based expression; Tables 1, 2). Plate transformants of BL21(DE3) pLysS to TYE agar containing 200 μg/ml ampicillin (40 μg/ml kanamycin for plasmid pβ′820–1407) and 35 μg/ml chloramphenicol, and incubate 12 h at 37°C. Plate transformants of XL1-blue to TYE agar containing 200 μg/ml ampicillin (40 μg/ml kanamycin for plasmid pβ1–235) and 20 μg/ml tetracycline, and incubate 16 h at 37°C.
Inoculate single colony into 5 ml LB containing antibiotics at concentrations specified in Step 1 in 15 ml culture tube with stainless steel closure, and shake vigorously for 12 h at 37°C. Transfer to 15 ml polypropylene centrifuge tube, and centrifuge 5 min at 3,000 × g at room temperature. Discard supernatant, wash cell pellet twice with 5 ml LB, and re-suspend cell pellet in 5 ml LB.
Inoculate into 1 L LB containing antibiotics at concentrations specified in Step 1 in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 1 ml 1 M IPTG, and shake vigorously for an additional 3 h [transformants of BL21(DE3) pLysS] or 5 h (transformants of XL1-blue) at 37°C.
Transfer culture to 1 L polypropylene copolymer centrifuge bottle. Harvest cells by centrifugation 20 min at 5,000 × g at 4°C.
Resuspend cell pellet in 10 ml buffer C containing 0.2% sodium desoxycholate and 0.02% lysozyme in 30 ml polypropylene copolymer centrifuge tube at 4°C. Place tube on ice. Lyse cells with five 30-s sonication pulses at 25% maximum sonicator output (2 min pause between each pulse).
Centrifuge 20 min at 15,000 × g at 4°C. Discard supernatant.
Resuspend pellet in 10 ml buffer C containing 0.2% n-octyl-β-D-glucopyranoside (0.5% Triton X-100 for preparation of σ70) and 0.02% lysozyme at 4°C. Sonicate as in Step 5. Centrifuge 20 min at 15,000 × g at 4°C. Discard supernatant.
Resuspend pellet in 10 ml buffer C containing 0.2% n-octyl-β-D-glucopyranoside (0.5% Triton X-100 for preparation of σ70) at 4°C. Sonicate as in Step 5. Centrifuge 20 min at 15,000 × g at 4°C. Discard supernatant.
Resuspend pellet in 10 ml buffer C at 4°C. Place tube on ice, and sonicate 10 s at 25% maximum sonicator output. Divide into 500 μl aliquots, and transfer to 1.5 siliconized polypropylene microcentrifuge tubes. Centrifuge 5 min at 13,000 × g at 4°C. Discard supernatant.
Add 100 μl ice-cold buffer C containing 10% glycerol to each aliquot. Store at −80°C (stable for at least 2 y). Expected yield: 50–100 mg (1.5–3 mg/aliquot). Expected purity: 50–90%.
3.3.3. Reconstitution of RNAP and RNAP derivatives
Thaw aliquots containing purified α subunit (from Section 3.3.1, Step 15) and crude recombinant RNAP subunits and subunit fragments (from Section 3.3.2, Step 10) by placing on ice for 10 min. Centrifuge 30 s at 13,000 × g at 4°C. Discard supernatants.
Resuspend each pellet in 500 μl buffer D. Incubate 30 min at 4°C, rocking gently. Centrifuge 5 min at 13,000 × g at 4°C.
Transfer supernatant to new 1.5 ml siliconized polypropylene microcentrifuge tubes at 4°C. Determine protein concentrations using Bio-Rad Protein Assay according to supplier’s protocol (expected concentrations: 3–6 mg/ml).
Prepare core reconstitution mixture by combining in 1.5 ml siliconized polypropylene microcentrifuge tube 30 μg N-terminally hexahistidine-tagged α, 300 μg β (or 170 μg β1–235 and 800 μg β235–1342; or 700 μg β1–989 and 300 μg β950–1342), 500 μg β′ (or 400 μg β′1–577 and 500 μg β′544–1407; or 700 μg β′1–877 and 330 μg β′820–1407), and 70 μg ω, and diluting with buffer D to a total protein concentration 450 μg/ml.
Prepare σ70 reconstitution mixture by adding 250 μg σ70 to 1.5 ml siliconized polypropylene microcentrifuge tube and diluting with buffer D to a total protein concentration 1500 μg/ml.
Dialyze core and σ70 reconstitution mixtures separately in collodion dialysis bags against two 1 L changes of buffer E for 16 h at 4°C.
Transfer core and σ70 reconstitution mixtures to separate 2.0 ml siliconized polypropylene microcentrifuge tubes. Centrifuge 5 min at 13,000 × g at 4°C. Combine supernatants in a single, new 2.0 ml siliconized polypropylene microcentrifuge tube.
Incubate 45 min at 30°C. Centrifuge 10 min at 13,000 × g at 4°C.
3.3.4. Purification of RNAP and RNAP derivatives
During incubation of Step 8 of Section 3.3.3, place 200 μl Ni:NTA-agarose in 2.0 ml siliconized polypropylene microcentrifuge tube, and centrifuge 2 min at 13,000 × g at 4°C. Remove supernatant.
Resuspend Ni:NTA-agarose in 1 ml buffer F containing 5 mM imidazole at 4°C. Centrifuge 2 min at 13,000 × g at 4°C. Remove supernatant. Repeat two times.
Add supernatant of Step 8 of Section 3.3.3 to Ni:NTA-agarose from Step 2. Incubate 45 min at 4°C, rocking gently. Centrifuge 2 min at 13,000 × g at 4°C. Discard supernatant.
Resuspend in 1.5 ml buffer F containing 5 mM imidazole at 4°C. Rock gently 15 s at 4°C. Centrifuge 2 min at 13,000 × g at 4°C. Discard supernatant. Repeat two times.
Resuspend in 250 μl buffer F containing 150 mM imidazole. Rock gently 2 min at 4°C. Centrifuge 2 min at 13,000 × g at 4°C.
Transfer supernatant to Nanosep-30K centrifugal concentrator. Centrifuge at 13,000 × g at 4°C until sample volume is reduced to ~50 μl (~15 min).
Transfer sample to 1.5 siliconized polypropylene microcentrifuge tube. Add, in order, 1 μl 0.1 M β-mercaptoethanol and 50 μl glycerol; mix well; and store at −20°C (stable for at least 1 month).
Determine protein concentration using Bio-Rad Protein Assay according to supplier’s protocol. Expected yield: 100 μg. Expected purity: > 90%.
3.4. Static photocrosslinking
3.4.1. Synthesis of N,N′-bisacryloylcystamine (BAC) (see Note 10)
Acryloyl chloride is highly toxic. Therefore, all manipulations in this section must be performed in a fume hood.
Dissolve 4.0 g (18 mmol) cystamine dihydrochloride in 40 ml 3 M NaOH (120 mmol). Dissolve 4.3 ml (54 mmol) acryloyl chloride in 40 ml chloroform. Mix solutions in 500 ml flask (see Note 11). (Two phases will form: an upper, aqueous phase; and a lower, organic phase.) Place flask on plate stirrer, and stir 3 min at room temperature, followed by 15 min at 50°C.
Discontinue stirring. Immediately transfer reaction mixture to 500 ml separating funnel, allow phases to separate (~2 min), and transfer lower, organic phase to 250 ml beaker.
Place on ice for 10 min. Collect precipitate by filtration in Büchner funnel.
Transfer precipitate to 250 ml beaker with 30 ml chloroform at room temperature. Place beaker on plate stirrer, and stir 1 min at room temperature, followed by 5 min at 50°C. Place on ice for 10 min, and collect precipitate (BAC) by filtration in Buchner funnel.
Transfer precipitate to 50 ml polypropylene centrifuge tube. Seal tube with Parafilm, pierce seal several times with syringe needle, place tube in vacuum desiccator, and dry under vacuum 16 h at room temperature. Expected yield: 1.5–1.9 g.
3.4.2. Preparation of polyacrylamide:BAC gel
Prepare 20% acrylamide:BAC (19:1) stock solution by dissolving, in order, 19 g acrylamide and 1g BAC in 80 ml water in 200 ml beaker at room temperature. Place on plate stirrer and stir 10 min at 60°C (see Note 12). Adjust volume to 100 ml with water. Allow solution to cool to room temperature. Filter stock solution using 0.22 μm filter unit, and store at room temperature in the dark (stable for at least 2 months).
Mix 9 ml 20% acrylamide:BAC (19:1) stock solution, 1.8 ml 10x TBE, and 25.2 ml water. Add 180μl TEMED and 90 μl freshly prepared 10% ammonium persulfate (see Note 13). Immediately pour into slab gel assembly with siliconized notched glass plate (27 × 16 × 0.1 cm) (see Note 14). Insert comb and heat slab gel assembly to ~60°C by positioning task lamp with 60 W tungsten bulb 2 cm from the outer glass plate (see Note 15). Allow 10–20 min for polymerization. (The polyacrylamide:BAC gel is stable for up to 72 h at 4°C.)
3.4.3. Formation and isolation of RNAP-promoter open complex [all steps carried out under subdued lighting (see Note 4)]
Place polyacrylamide:BAC slab gel in electrophoresis apparatus, clip 10 × 25 cm silicone heating mat to directly to outer glass plate of the slab gel assembly with four large binder clips, and pour 0.5x TBE buffer in upper and lower reservoirs.
Pre-run gel for 2 h at 20 V/cm.
Pre-warm electrophoresis unit by placing in 37°C cabinet for 3 h.
During 37°C pre-warming of Step 3, dilute RNAP or RNAP derivative to 180 μg/ml (400 nM) in buffer F containing 1 mM β-mercaptoethanol and 50% glycerol.
Immediately after 37°C pre-warming of Step 3, add the following, in order, to a 1.5 ml siliconized polypropylene microcentrifuge tube: 2 μl 5 nM derivatized DNA fragment (~5,000 Ci/mmol), 5 μl 2x DTT-free transcription buffer, 2 μl water and 1 μl 180 μg/ml (400 nM) RNAP or RNAP derivative (all at room temperature).
Incubate 20 min at 37°C in the dark.
During incubation of Step 6 apply voltage to gel: 16 V/cm. Wash wells of gel carefully with 0.5x TBE to remove unpolymerized acrylamide and BAC. (Caution: Care must be exercised to avoid electrocution.) Connect heating mat to variable voltage controller. Monitor gel temperature at 5 min intervals by inserting thermocouple probe into the gel for 5 s (and removing immediately thereafter). Maintain gel temperature at 37°C, adjusting heater voltage as necessary (typically 12–14V).
After completing incubation of Step 6, immediately add 1 μl 0.22 μg/ml heparin (pre-warmed to 37°C), mix, and immediately apply sample to gel (see Note 16). Load 5 μl nondenaturing loading buffer into adjacent lane. (Caution: Care must be exercised to avoid electrocution.) Continue electrophoresis 20 min at 16 V/cm. Monitor gel temperature at 5 min intervals by inserting thermocouple probe into the gel for 5 s (and removing immediately thereafter). Maintain gel temperature at 37°C, adjusting heater voltage as necessary (typically 12–14V).
Immediately proceed to next step (Section 3.4.4).
3.4.4. UV-irradiation of RNAP-promoter open complex
Remove gel with both glass plates in place (see Note 17), and mount vertically in a Rayonet RPR-100 photochemical reactor equipped with 16 RPR-3500 Å tubes.
Immediately UV-irradiate 3 min (17 mJ/mm2 at 350 nm) (see Note 18). (Caution: Care must be exercised to avoid injury to eyes and skin.)
Immediately proceed to next step (Section 3.4.5).
3.4.5. Identification, excision, and solubilization of portion of gel containing RNAP-promoter open complex
Remove one glass plate, and cover gel with plastic wrap (leaving other glass plate in place). Attach two autoradiography markers. Expose to x-ray film for 1.5 h at room temperature (see Note 19). Process film.
Cut out portion of film corresponding to RNAP-promoter complex of interest. Using light box, superimpose cut-out film on gel, using autoradiography markers as reference points. Using disposable scalpel, excise portion of gel corresponding to RNAP-promoter complex. Transfer excised gel slice to 1.5 ml siliconized microcentrifuge tube.
Solubilize gel slice by adding 10 μl 1 M DTT (~0.4 M final), and heating 5 min at 37°C (see Note 20).
Immediately proceed to next step (Section 3.4.6).
3.4.6. Nuclease digestion
During x-ray film exposure of Step 1, Section 3.4.5, dilute DNaseI and micrococcal nuclease with ice-cold nuclease dilution solution to final concentration 10 u/μl.
Transfer 10 μl to new 1.5 ml siliconized polypropylene microcentrifuge tube, and add 1 μl 200 mM CaCl2, 1 μl 0.1 mM PMSF, 0.5 μl (5 units) micrococcal nuclease and 0.5 μl (5 units) DNase I. Incubate 20 min at 37°C. Terminate reaction by adding 3 μl 5x SDS loading buffer, and heating 5 min at 100°C.
Immediately proceed to next step (Section 3.4.7).
3.4.7. Analysis
Apply entire sample (16 μl) to 7–15% gradient polyacrylamide (37.5:1 acrylamide:bisacrylamide) slab gel. As marker, load into adjacent lane 5 μl prestained protein molecular weight markers. Electrophorese in SDS running buffer at 25 V/cm until bromophenol blue reaches bottom of gel.
Dry gel, and autoradiograph or perform storage-phosphor imaging.
Identify bands corresponding to crosslinked proteins.
3.5. Kinetic photocrosslinking
3.5.1 Rapid-quench-flow mixing and pulsed-laser UV-irradiation [steps 15–26 carried out under subdued lighting (see Note 4)]
Start laser, and allow laser to operate for 1 h. Open 355 nm laser port, and check laser pulse energy using laser calorimeter and laser energy meter. Adjust laser pulse energy to 80 mJ/cm (a pulse energy that minimizes damage to optical windows of Raman cell and sample chamber). (Caution: Care must be exercised to avoid injury. All contact with the laser beam must be avoided. All work with laser must be performed by persons having laser-safety training and wearing appropriate protective gear, including appropriate laser safety goggles, laboratory coat, and gloves. Persons not having laser-safety training and/or not having appropriate protective gear must be excluded from the room. The room must be windowless or have fully blocked windows. The entrance to the room must have a warning light that operates when the laser operates and must have a supply of appropriate laser goggles, for all wavelengths and all energies used.)
Connect circulating water bath to rapid-quench-flow system, and allow to operate at 37°C for 1 h
Start rapid-quench-flow-system controller, and allow to operate for 10 min.
Using a nail, puncture hole in lid of each of 16 1.5 ml siliconized polypropylene microcentrifuge tubes (in order to permit insertion of exit line rapid-quench-flow system through lid, allowing collection of sample without splashing).
Add 11 ml of DTT- and glycerol-free transcription buffer to a 50 ml polypropylene centrifuge tube, and equilibrate at 37°C using a water bath. Add and dissolve 1.1 mg BSA (yielding DTT- and glycerol-free, BSA-containing transcription buffer).
Add 5 ml of urea/NaI solution to a 50 ml polypropylene centrifuge tube. Add 176 μl 14.2 M β-mercaptoethanol (yielding quench solution; see Note 23). Equilibrate at 37°C using a water bath.
Set the left and right syringe-load valves of the rapid-quench-flow system to the load position. Using two Luer-Lok 5 ml disposable syringes, load the two drive syringes of the rapid-quench-flow system with DTT- and glycerol-free, BSA-containing transcription buffer. (To remove air bubbles from the drive syringes while loading, work the solution back and forth rapidly several times.)
Set the center syringe-load valve of the rapid-quench-flow system to the load position. Using a Luer-Lok 5 ml disposable syringe, load the third drive syringe of the quench-flow device with 5 ml of the quench solution.
Set the center syringe-load valve of the rapid-quench-flow system to connect the loading and firing lines. Fill Luer-Lok 50 μl Hamilton syringe with the quench solution; attach syringe to loading port of the third drive syringe of the rapid-quench-flow system; and flush connection lines, flow cell, and exit line.
Set all valves of the rapid-quench-flow system to the fire position. Set the rapid-quench-flow-system drive plate to the ready position.
Set the sample-load valves of the rapid-quench-flow system to the flush position. Flush sample loops, flow cell, and exit line with 1 ml of methanol and then with 1 ml of water. Dry flushed areas for 1 min using compressed argon.
Using the Linagraph direct-print paper, verify that the laser beam passes through the optical windows of the sample chamber. Adjust optics, if necessary. Using the laser calorimeter and laser energy meter, quantify laser pulse energy at front optical window of the sample chamber. If necessary, adjust laser pulse energy to 1 mJ.
Establish electrical connections between the digital delay/pulse generator and the power supplies of the laser and the rapid-quench-flow system.
Prepare 40 nM solution of RNAP in 170 μl of DTT- and glycerol-free, BSA-containing transcription buffer (see Note 24). Fill a 1 ml tuberculin-slip-tip disposable syringe with the resulting solution.
Prepare 1 nM solution of derivatized promoter DNA fragment in 170 μl of DTT- and glycerol-free, BSA-containing transcription buffer (see Note 24). Fill a 1 ml tuberculin slip tip disposable syringe with the resulting solution.
Set the sample-load valves of the rapid-quench-flow system to the sample-load position. Attach syringes with RNAP and DNA solutions to the sample-load lines. Fill each load line to the tubing connector of the sample-load valve.
Program the rapid-quench-flow-system controller, entering the following settings: pre-flash volume, 40 μl; post-flash volume, 240 μl; delay time after flash, 0; and pre-flash reaction time, x1 (where x1 is the desired time, in seconds, between rapid-quench-flow mixing and pulsed-laser UV-irradiation).
Program the digital delay/pulse generator, entering the following setting: delay time, x1 (where x1 is the desired time, in seconds, between rapid-quench-flow mixing and pulsed-laser UV-irradiation).
Perform test firing. Press “G” button on the rapid-quench-flow-system controller to enable system. Press “fire” button on the laser controller to fire.
Flush and dry the sample loops, the flow cell, and the exit line as in step 11.
Using the 50 μl Hamilton syringe connected to the loading port of the third drive syringe in step 9, draw 20 μl of quenching solution out of the flow-cell connection tube.
Turn the sample-load valve to the sample-load position, and load the RNAP and DNA samples, one at a time, into the sample loops, bringing the meniscus of each sample to the 10 μl mark on each sample loop. (Sample loops should be pre-calibrated and pre-marked.)
Insert the exit line through the hole in the lid of a 1.5 ml siliconized polypropylene microcentrifuge tube from step 4.
Fire system (as described for test firing in step 19). Remove the collection tube with the expelled sample (~140 μl).
Repeat steps 16–18 and 20–23 to perform 16 separate experiments, each with a different pre-flash reaction time, x2 through x16, on the rapid-quench-flow system and delay time, x2 through x16, on the delay/pulse generator (see Note 25).
Immediately proceed to the next step (section 3.5.2).
3.5.2. Nuclease digestion
Dilute DNase I, and micrococcal nuclease with ice-cold nuclease dilution solution to a final concentration of 50 units/μl.
Dilute OmniCleave endonuclease with ice-cold dilution buffer to a final concentration of 50 units/μl.
Prepare digestion mixture, containing: 20 μl of 200 mM CaCl2, 20 μl of 200 mM MgCl2, 4 μl of the solution from step 1, and 4 μl of the solution from step 2.
Transfer 34 μl of each sample from section 3.5.1 to a new 1.5-ml siliconized polypropylene microcentrifuge tube, and add 2 μl of the digestion mixture from step 2. Incubate for 15 min at 37°C. Terminate reaction by adding 9 μl of 5X SDS loading buffer and heating for 5 min at 100°C.
Immediately proceed to the next step (section 3.5.3).
3.5.3. Analysis
Apply the entire sample (45 μl) to a Criterion precast 4–20% Tris-HCl gradient slab gel. As a marker, load 5 μl of Precision-Plus protein dual-color standards into the adjacent lane. Electrophorese in SDS running buffer at 25 V/cm until the bromophenol blue is ~0.5 cm from the bottom of the gel.
Dry gel, and perform storage-phosphor imaging.
Identify bands corresponding to crosslinked proteins.
Quantify intensities of bands corresponding to crosslinked proteins.
Plot intensities vs. time, using data from at least three independent determinations.
-
Perform curve fitting to extract kinetic parameters as follows:
For crosslinked species for which intensities exhibit a monoexponential rise to a maximum, fit data to:
where y is intensity, t is the time in seconds between rapid-quench-flow mixing and pulsed-laser UV-irradiation DNA and RNAP, kobs is the pseudo-first-order rate constant C0 is the intensity at t =0, and A is the intensity at t = ∝.For crosslinked species for which intensities exhibit a biexponential rise to a maximum, fit data to:
where y is intensity, t is the time in seconds between rapid-quench-flow mixing and pulsed-laser UV-irradiation DNA and RNAP, k1,obs and k2,obs are pseudo-first-order rate constants for process 1, and process 2, and where A1 and A2 are the intensities at t = ∝ of process 1 and process 2.
4. Notes
M13mp2(ICAP-UV5) carries the lacP(ICAP-UV5) promoter, a derivative of the lacP promoter having a consensus DNA site for CAP and a consensus −10 element (5). M13mp2(ICAP-UV5)-rev carries the lacP(ICAP-UV5) promoter in an orientation opposite to that in M13mp2(ICAP-UV5) (5). M13mp2(ICAP-UV5)-rev was prepared from M13mp2(ICAP-UV5) by deleting the PvuII-PvuII segment corresponding to positions −217 to −125 of lacP(ICAP-UV5) and inverting the PvuII-PvuII segment corresponding to positions −124 to +145 of lacP(ICAP-UV5).
M13mp2(ICAP-UV5) and M13mp2(ICAP-UV5)-rev ssDNAs carry, respectively, the non-template and template strands of lacP(ICAP-UV5). M13mp2(ICAP-UV5) and M13mp2(ICAP-UV5)-rev ssDNAs are prepared as in 10.
The specified 10x digestion buffer is for PvuII and HaeIII. Use 10x digestion buffer recommended by supplier--omitting DTT (see 44)--for other restriction enzymes.
Fluorescent light and daylight must be excluded. Low to moderate levels of incandescent light (e.g., from single task lamp with 60 W tungsten bulb) are acceptable.
The derivatized oligodeoxyribonucleotide tolerates exposure to the Hitachi L-3000 diode-array HPLC UV detector. The derivatized oligodeoxyribonucleotide can be identified unambiguously by monitoring the UV-absorbance spectrum from 200 nm to 350 nm. The derivatized oligodeoxyribonucleotide exhibits an absorbance peak at 260 nm, attributable to DNA, and a shoulder at 300–310 nm, attributable to the azidophenacyl group.
The derivatization procedure yields two diastereomers in an approximately 1-to-1 ratio: one in which azidophenacyl is incorporated at the sulfur atom corresponding to the phosphate O1P, and one in which azidophenacyl is incorporated at the sulfur atom corresponding to the phosphate O2P (see 7,8). Depending on oligodeoxyribonucleotide sequence and HPLC conditions, the two diastereomers may elute as a single peak, or as two peaks (e.g., at 24% and 25% solution B). In most cases, no effort is made to resolve the two diastereomers, and experiments are performed using the unresolved diastereomeric mixture. This permits simultaneous probing of protein-DNA interactions in the DNA minor groove (probed by the O1P-derivatized diastereomer) and the DNA major groove (probed by the O2P-derivatized diastereomer).
Phenyl azides are unstable at temperatures above 70°C. Avoid heating above 70°C.
HaeIII digestion, which yields a DNA fragment corresponding to positions −141 to +63 of lacP(ICAP-UV5), is used for preparation of DNA fragments derivatized between positions −80 and −1, inclusive. PvuII digestion, which yields a DNA fragment corresponding to positions −124 to +145 of lacP(ICAP-UV5), is used for preparation of DNA fragments derivatized between positions +1 and +80 inclusive. [Use of DNA fragments with >60 bp between the site of derivatization and the nearest DNA-fragment end eliminates “non-specific” crosslinking from the sub-population of complexes having RNAP bound at a DNA-fragment end (45) rather than at the promoter.]
The procedures described are for “in-gel” static photocrosslinking, in which complexes are prepared in solution, isolated by non-denaturing polyacrylamide gel electrophoresis, and UV-irradiated in situ in the polyacrylamide gel matrix; 2,5,6). In-gel procedures facilitate preparation, isolation, and analysis of homogeneous complexes. For alternative procedures, involving “on-bead” static photocrosslinking, see 4. For alternative procedures, involving “in-solution” static photocrosslinking, see 1,5.
BAC is a disulfide-containing analog of bisacrylamide (46–48). Polyacrylamide:BAC gels can be solubilized by addition of reducing agents (46–48). The synthesis of BAC in this chapter is adapted from 46.
Acryloyl chloride reacts violently with water. Add acryloyl chloride in 0.5 ml portions, waiting 30 s between successive additions.
BAC is substituted for bisacrylamide on a mole-equivalent, not mass-equivalent, basis (46–48). The solubility of BAC in water is increased by adding acrylamide before adding BAC, and by performing additions at 60°C.
TEMED and ammonium persulfate concentrations are critical variables in preparation of polyacrylamide:BAC gels (47,48). (Use of non-optimal TEMED and ammonium persulfate concentrations in preparation of polyacrylamide:BAC results in difficulties in subsequently solubilizing gels.)
Siliconize notched glass plate by applying 30 μl SurfaSil siliconizing agent and spreading evenly with Kimwipe.
Heating during polymerization yields polyacrylamide:BAC gels that are maximally solubilizable upon addition of reducing agents (47,48). Heat the glass plates of the gel assembly evenly. (If necessary, use two task lamps.) Avoid heating above 70°C, as this can result in formation of bubbles and/or detachment of gels from the glass plates.
Do not add loading buffer to the reaction mixture. The reaction mixture is sufficiently dense for loading (due to the presence of glycerol).
UV-irradiation is performed with both glass plates in place. The glass plates exclude wavelengths <300 nm, minimizing photodamage to protein and DNA. It is important to verify that the plates exhibit absorbances of 1.5 AU at 320 nm (e.g., by sacrificing a glass plate and placing a piece in the cuvette holder of a UV/Vis spectrophotometer). Glass plates purchased from Aladin, Inc. (Aladin Enterprises, Inc., 1255 23rd Avenue, San Francisco CA 94122 USA) have performed satisfactorily.
For in-gel UV-irradiation of RNAP-promoter intermediate complex, pre-chill photochemical reactor 15 min in 15°C cabinet.
Do not use tight-fitting X-ray autoradiography cassettes, which can squeeze and distort the gel on the glass plate during exposure. The Kodak X-ray exposure holder with intensifying screen has performed satisfactorily.
2–4 M β-mercaptoethanol can be substituted for 1 M DTT.
For rapid mixing, we use a commercial microprocessor-controlled, stepping-motor-driven rapid-quench-flow system modified to incorporate a 20 μl sample chamber with OP1 acrylic optical windows (KinTek RQF-3 with modifications; see 49,50; Figure 2). The dead time for mixing is ~10 ms.
For laser UV-irradiation, we use a nanosecond pulsed Nd-YAG laser with third harmonic generator (Continuum Powerlite 7010-DS/TS/QS), a Raman cell (Light-Age RC-1), a Pellin-Broca prism, 355 nm laser optics (first mirror) and 309 nm laser optics (all other optics) (see 51,52; Figure 2). This system is able to generate up to 5 mJ per 6 ns pulse at 309 nm (a wavelength selected to be close to the absorbance maximum of the azidophenacyl photoactivatible crosslinking agent and well separated from the absorbance maxima of DNA and protein).
Laser UV-irradiation of phenyl-azide photoactivatible crosslinking agents reportedly produces only short-lived reactive species (lifetimes ~5 ms in heptane; lifetimes ≪1 ms in the presence of nucleophiles; 53–55). Nevertheless, to ensure effectively immediate inactivation of reactive species and effectively immediate termination of crosslinking, we quench reactions immediately after laser UV-irradiation by mixing with a quench solution of 5 M urea, 0.5 M NaI, and 0.5 M β-mercaptoethanol. The dead time for quenching is ~10 ms.
Use of a 40-fold mole excess of RNAP over promoter DNA, ensures that kinetics of RNAP-DNA association are pseudo-first order.
Each set of 16 experiments requires ~1 h. When performing successive sets of experiments, freshly made solutions should be prepared for each set of experiments.
Acknowledgments
The basic protocol for preparation of derivatized DNA fragments was developed by T. Lagrange, the basic protocol for preparation of RNAP was developed by H. Tang and K. Severinov, and the basic protocol for in-gel static photocrosslinking was developed by T.-K. Kim. We thank K. Severinov for plasmids; T.-K. Kim, T. Lagrange, D. Reinberg, and K. Severinov for discussion; and a Howard Hughes Medical Institute Investigatorship and National Institutes of Health grant GM41376 to R.H.E. for financial support.
References
- 1.Lagrange T, Kim TK, Orphanides G, Ebright Y, Ebright RH, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: Organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim TK, Lagrange T, Wang YH, Griffith J, Reinberg D, Ebright RH. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagrange T, Kapanidis A, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 5.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 6.Kim T-K, Lagrange T, Naryshkin N, Reinberg D, Ebright RH. Site-specific protein-DNA photocrosslinking. In: Travers A, Buckle M, editors. Protein-DNA Interactions: A Practical Approach. IRL Press; Oxford: 2000. pp. 319–335. [Google Scholar]
- 7.Fidanza J, Ozaki H, McLaughlin L. Site-specific labeling of DNA sequences containing phosphorothioate diesters. J Amer Chem Soc. 1992;114:5509–5517. [Google Scholar]
- 8.Yang SW, Nash H. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer A, Barany F. Photoaffinity cross-linking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene. 1995;153:1–8. doi: 10.1016/0378-1119(94)00752-e. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 11.Wang Y, Stumph W. Identification and topological arrangement of Drosophila proximal sequence element (PSE)-binding protein subunits that contact the PSEs of U1 and U6 small nuclear RNA genes. Mol Cell Biol. 1998;18:1570–1579. doi: 10.1128/mcb.18.3.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett M, Thomm M, Geiduschek E. The orientation of DNA in an archaeal transcription initiation complex. Nature Struct Biol. 2000;7:782–785. doi: 10.1038/79020. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett M, Thomm M, Geiduschek E. Topography of the euryarchaeal transcription initiation complex. J Biol Chem. 2004;279:5894–5903. doi: 10.1074/jbc.M311429200. [DOI] [PubMed] [Google Scholar]
- 14.Renfrow M, Naryshkin N, Lewis M, Chen HT, Ebright RH, Scott R. Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J Biol Chem. 2004;279:2825–2831. doi: 10.1074/jbc.M311433200. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, Mandal S, Hampsey M. High-resolution protein-DNA contacts for the yeast RNA polymerase II general transcription machinery. Biochem. 2004;43:12741–12749. doi: 10.1021/bi048993r. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, Geiger J, Hahn S, Sigler P. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Nikolov D, Burley S. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 18.Geiger J, Hahn S, Lee S, Sigler P. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 19.Tan S, Hunziker Y, Sargent D, Richmond T. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 20.Nikolov D, Chen H, Halay E, Usheva A, Hisatake K, Lee DK, Roeder R, Burley S. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 21.Bartholomew B, Kassavetis G, Braun B, Geiduschek EP. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartholomew B, Kassavetis G, Geiduschek EP. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun B, Bartholomew B, Kassavetis G, Geiduschek EP. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 24.Kassavetis G, Kumar A, Ramirez E, Geiduschek EP. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol Cell Biol. 1998;18:5587–5599. doi: 10.1128/mcb.18.9.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 26.Coulombe B, Li J, Greenblatt J. Topological localization of the human transcription factors IIA, IIB, TATA Box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 27.Gong X, Radebaugh C, Geiss G, Simon S, Paule M. Site-directed photo-cross-linking of rRNA transcription initiation complexes. Mol Cell Biol. 1995;15:4956–4963. doi: 10.1128/mcb.15.9.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis E, Wolffe A. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 29.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 30.Forget D, Robert F, Grondin G, Burton Z, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Record MT, Reznikoff W, Craig M, McQuade K, Schlax P. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhart F, editor. Escherichia coli and Salmonella. Vol. 1. ASM Press; Washington, D.C: 1996. pp. 792–820. [Google Scholar]
- 32.Ebright R. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 33.Severinov K, Mustaev A, Severinova E, Bass I, Kashlev M, Landick R, Nikiforov V, Goldfarb A, Darst S. Assembly of functional Escherichia coli RNA polymerase containing β subunit fragments. Proc Natl Acad Sci USA. 1995;92:4591–4595. doi: 10.1073/pnas.92.10.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst S, Goldfarb A. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the β and β′ subunits of Escherichia coli RNA polymerase. J Biol Chem. 1996;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 35.Tang H, Severinov K, Goldfarb A, Ebright RH. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H, Kim Y, Severinov K, Goldfarb A, Ebright RH. Escherichia coli RNA polymerase holoenzyme: rapid reconstitution from recombinant α, β, β′, and σ subunits. Meths Enzymol. 1996;273:130–134. doi: 10.1016/s0076-6879(96)73012-4. [DOI] [PubMed] [Google Scholar]
- 37.Martin E, Sagitov V, Burova E, Nikiforov V, Goldfarb A. Genetic dissection of the transcription cycle. A mutant RNA polymerase that cannot hold onto a promoter. J Biol Chem. 1992;267:20175–20180. [PubMed] [Google Scholar]
- 38.Zalenskaya K, Lee J, Chandrasekhar NG, Shin YK, Slutsky M, Goldfarb A. Recombinant RNA polymerase: inducible overexpression, purification and assembly of Escherichia coli rpo gene products. Gene. 1990;89:7–12. doi: 10.1016/0378-1119(90)90199-2. [DOI] [PubMed] [Google Scholar]
- 39.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis A, Niu W, Ebright Y, Levy R, Ebright R. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 40.Ebright RH, Ebright Y, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucl Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert W. Starting and stopping sequences for the RNA polymerase. In: Losick R, Chamberlin M, editors. RNA Polymerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1976. pp. 193–206. [Google Scholar]
- 42.Zhang X, Zhou Y, Ebright Y, Ebright RH. Catabolite gene activator protein (CAP) is not an acidic-activating-region transcription activator protein: negatively charged amino acids of CAP that are solvent-accessible in the CAP-DNA complex play no role in transcription activation at the lac promoter. J Biol Chem. 1992;267:8136–8139. [PubMed] [Google Scholar]
- 43.Kunkel T. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staros J, Bayley H, Standring D, Knowles J. Reduction of aryl azides by thiols: implications for the use of photoaffinity reagents. Biochem Biophys Res Commun. 1978;80:568–572. doi: 10.1016/0006-291x(78)91606-6. [DOI] [PubMed] [Google Scholar]
- 45.Melancon P, Burgess R, Record M. Direct evidence for the preferential binding of Escherichia coli RNA polymerase holoenzyme to the ends of deoxyribonucleic acid restriction fragments. Biochem. 1983;22:5169–5176. doi: 10.1021/bi00291a017. [DOI] [PubMed] [Google Scholar]
- 46.Hansen JN. Electrophoresis of ribonucleic acid on a polyacrylamide gel which contains disulfide cross-linkages. Anal Biochem. 1976;76:37–44. doi: 10.1016/0003-2697(76)90261-x. [DOI] [PubMed] [Google Scholar]
- 47.Hansen JN. Chemical and electrophoretic properties of solubilizable disulfide gels. Anal Biochem. 1980;105:192–201. doi: 10.1016/0003-2697(80)90445-5. [DOI] [PubMed] [Google Scholar]
- 48.Hansen JN. Use of solubilizable acrylamide disulfide gels for isolation of DNA fragments suitable for sequence analysis. Anal Biochem. 1981;116:146–151. doi: 10.1016/0003-2697(81)90337-7. [DOI] [PubMed] [Google Scholar]
- 49.Langowski J, Urbanke C, Scuppe E. Construction of a microprocessor-controlled pulsed quench-flow apparatus for the study of fast chemical and biochemical reactions. Anal Biochem. 1984;142:91–97. doi: 10.1016/0003-2697(84)90521-9. [DOI] [PubMed] [Google Scholar]
- 50.Johnson K. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Meths Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 51.Hockensmith J, Kubasek W, Vorachek W, von Hippel P. Laser cross-linking of nucleic acids to proteins: methodology and first application to the phage T4 DNA replication system. J Biol Chem. 1986;261:3512–3518. [PubMed] [Google Scholar]
- 52.Hockensmith J, Kubasek W, Vorachek W, Evertsz E, von Hippel P. Laser cross-linking of protein-nucleic acid complexes. Meths Enzymol. 1991;208:211–236. doi: 10.1016/0076-6879(91)08015-a. [DOI] [PubMed] [Google Scholar]
- 53.Shields C, Chrisope D, Schuster G, Dixon A, Poliakoff M, Turner J. Photochemistry of aryl azides: detection and characterization of a dehydroazepine by time-resolved infrared spectroscopy and flash photolysis at room temperature. J Amer Chem Soc. 1987;109:4723–4726. [Google Scholar]
- 54.Li YZ, Kirby J, George M, Poliakoff M, Schuster G. 1,2-Didehydroazepines from the photolysis of substituted aryl azides: analysis of their chemical and physical properties by time-resolved spectroscopic methods. J Amer Chem Soc. 1988;110:8092–8098. [Google Scholar]
- 55.Marcinek A, Leyva E, Whitt D, Platz M. Evidence for stepwise nitrogen extrusion and ring expansion upon photolysis of phenyl azide. J Amer Chem Soc. 1993;115:8609–8612. [Google Scholar]