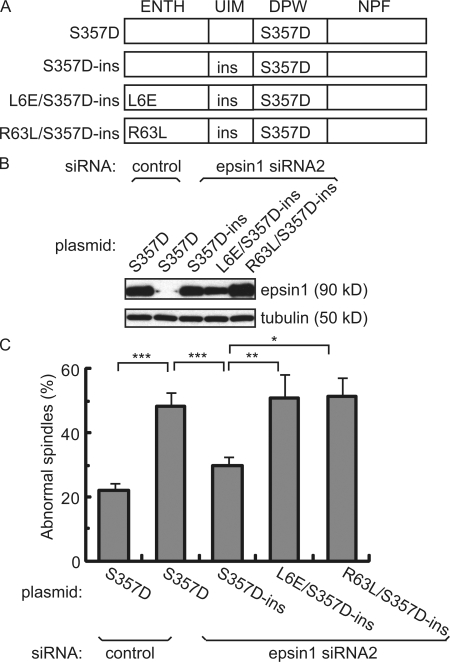
Figure 4.
Epsin1 regulates spindle integrity through its membrane-bending ENTH domain. (A) Domain structure of epsin1 expression constructs with the indicated mutations. The ENTH domain interacts with membrane and has membrane-bending activity. The ubiquitin-interacting motif (UIM) binds to ubiquitin. DPW motifs interact with AP2. The NPF motif interacts with proteins such as Eps15. The mutant epsin1Ser357D (S357D) mimics the mitotic phosphorylated form. The RNAi-insensitive rescue construct epsin1Ser3573D-ins (S357D-ins) contains mutations in wobble codons in the sequence corresponding to siRNA2, which do not affect protein sequences. Mutant epsins epsin1L6E/Ser357D-ins (L6E/S357D-ins) and epsin1R63L/Ser357D-ins (R63L/S357D-ins) are derived from epsin1Ser357D-ins but contain an additional point mutation in the ENTH domain that abolishes the membrane-bending activity of epsin1. (B) Western blot analysis of epsin1 expression in HeLa cells. Epsin1Ser357D expression was inhibited by siRNA2 but not by control siRNA, whereas expression of epsin1Ser357D-ins, epsin1L6E/Ser357D-ins, and epsinR63L/Ser357D-ins was not affected by treatment with siRNA2. (C) Rescue of spindle morphology defects in HeLa cells by epsin1Ser357D-ins but not by epsin1L6E/Ser357D-ins or epsin1R63L/Ser357D-ins. The percentage of defective spindles, as judged by α-tubulin and pericentrin staining, was quantified in HeLa cells cotransfected with the siRNA oligonucleotide and each expression construct. The percentages of cells with abnormal spindle morphology from at least three independent experiments are shown, with >100 prometaphase and metaphase cells counted in each condition per experiment. Error bars show standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001.