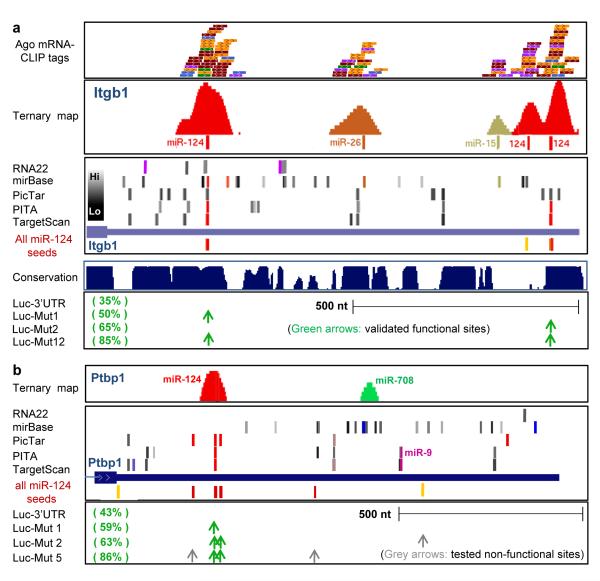
Fig. 3. Ago-miRNA ternary clusters in validated miR-124 mRNA targets.
(a) Ago—mRNA CLIP tags (top panel: raw tags, one color per biologic replicate: second panel: ternary map of Ago-mRNA normalized clusters around top predicted miRNA sites) compared with predicted miRNA sites (using indicated algorithms) for the 3′ UTR of Itgb1 (third panel, see Supplementary Methods; colors indicate predicted top 20 miRNAs as in Fig. 5; grey bars indicate miRNAs ranking below the top 20 (per heat-map) in Ago-miRNA CLIP). All predicted miR-124 6-8 mer seeds (conserved (red) or non-conserved (yellow) sites) are shown. Bottom panel shows data from luciferase assays in which mutagenesis of predicted miR-124 seeds at the indicated positions had the indicated effects on restoring from miR-124 mediated suppression (35% of baseline luciferase levels)41. (b) Ago-miRNA ternary maps compared with previously reported functional data for Ptbp142; see Supplementary Fig. 10 for maps of Ctdsp144 and Vamp344.