Fig. 4. Meta-analysis of Ago-mRNA clusters in large-scale screens of miR-124 regulated targets.
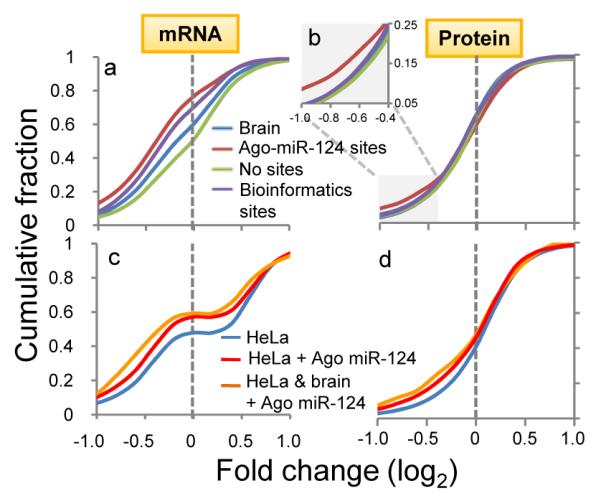
(a) Transcripts with predicted (conserved) miR-124 seeds (purple line) showed miR-124 suppression relative to all transcripts expressed in brain and cell lines (blue line) or those with no miR-124 seed sequences (green line). Transcripts with Ago-miR-124 ternary clusters (containing both miR-124 seed sequences and Ago-mRNA CLIP tags; red line) showed further miR-124 suppression. (b) Similar results were seen when analyzing miR-124 dependent protein suppression (identified by SILAC)11, with discrimination by the presence of Ago-miR-124 ternary clusters especially evident where there were smaller numbers of transcripts showing larger changes (log2<-0.4; inset). (c) Transcripts expressed in miR-124 transfected HeLa cells that harbor new Ago-miR-124 clusters (red line; or a subset of transcripts also harboring Ago-miR-124 clusters in mouse brain; yellow line), compared with previous analysis7 of regulated transcripts in miR-124 transfected HeLa cells. (d) As in (c), plotted for predicted protein levels, compared with prior data11.
