Abstract
Immunologic pathways involved in sarcoidosis pathogenesis are largely unknown. We hypothesized that patients with sarcoidosis have characteristic mRNA profiles. Microarray analysis of gene expression was done on peripheral blood (12 patients,12 controls), lung (6 patients, 6 controls) and lymph node (8 patients, 5 controls). Comparing peripheral blood from patients with sarcoidosis to controls, 872 transcripts were upregulated and 1039 were downregulated at ≥ 1.5-fold change and a significant q value. Several transcripts associated with interferon and STAT1 were upregulated. Lung and lymph node analyses also showed dramatic increases in STAT1 and STAT1-regulated chemokines. Granulomas in lymph nodes of patients with sarcoidosis expressed abundant STAT1 and phosphorylated STAT1. STAT1 might play an important role in sarcoidosis. This novel hypothesis unites seemingly disparate observations with regard to sarcoidosis including implication of a casual role for interferons, a suspected infectious trigger, TH1 predominating lymphocytes in bronchoalveolar lavage, and the association with hypercalcemia.
Keywords: Gene expression profiling, microarray analysis, sarcoidosis, uveitis
INTRODUCTION
Sarcoidosis is a granulomatous disease presumably induced by a harmful immune response. Sarcoidosis can be present in many organ systems including the lung, lymph node, eye, skin, joint, heart, liver, and brain. Although the trigger for the immunological damage is unknown, an infectious or environmental precipitant is strongly suspected [1]. Genetic factors also contribute substantially to the development of this disease [2].
Microarray-based assays have allowed the detection of thousands of mRNA transcripts from relatively small samples. The study of gene expression has been especially informative in the analysis of malignant tissue such as lymphoma [3], melanoma [4], or breast cancer [5]. In these examples, the pattern of gene expression provides diagnostic and prognostic information which cannot be obtained by histological analysis.
Microarray analysis has contributed to elucidating the pathogenesis of immune-mediated diseases. For example, many but not all patients with systemic lupus erythematosus (SLE) have an upregulation of genes induced by type I interferons [6]. Peripheral blood RNA also has distinct expression patterns in immune mediated diseases such as rheumatoid arthritis [7], multiple sclerosis [8], and dermatomyositis [9].
In order to derive clues about the pathogenesis of sarcoidosis, we performed microarray analysis of gene expression using peripheral blood samples from patients with sarcoidosis. We compared gene expression in peripheral blood with gene expression detected in either lung or lymph node from patients with this disease. Our study indicates that many genes under the regulation of the transcription factor, STAT1 (signal transducer and activator of transcription 1), have increased peripheral blood expression in sarcoidosis. The STATs are a family of transcription factors that regulate a set of genes involved in the inflammatory response [10]. STAT1, in particular, is induced by interferons that could be stimulated by viral or mycobacterial infection, potential triggers for sarcoidosis. Furthermore, we found that genes regulated by STAT1 were also upregulated in the lymph node from patients with sarcoidosis and that mRNA for at least 3 STAT1 regulated chemokines, CXCL9, CXCL10, and CXCL11, were markedly upregulated in either the lung or lymph node of patients with sarcoidosis. Finally, both STAT1 and the activated form of STAT1 (phosphorylated STAT1) were abundantly expressed in the lymph node granulomas of patients with sarcoidosis STAT1 may be a novel and promising target for pharmacotherapy of this disease since sarcoidosis is inconsistently responsive to immunosuppressive therapy [11].
METHODS
This study involved collaboration of two separate institutions which used slightly different microarray methodologies. All microarray studies on peripheral blood were performed at the Oregon Health & Science University while all solid tissue microarray studies were performed at Ohio State University Medical Center. The study received approval by the OHSU and OSUMC local institutional review boards for the blood and solid tissue studies respectively. Informed written consent was obtained from all patients and control subjects.
Human subject selection and diagnosis
Patients whose peripheral blood gene expression was analyzed had symmetric hilar adenopathy as judged either by chest x-ray or by computerized tomographic scan (CT). Since the combination of symmetric hilar adenopathy and uveitis is considered specific for a diagnosis of sarcoidosis [12], five of the seven patients with uveitis did not have biopsy confirmation of the diagnosis. Four of the five patients without uveitis had lung biopsy confirmation of the presence of non-caseating granuloma. The diagnosis of uveitis was confirmed on a dilated eye examination at a clinic which specializes in the care of patients with uveitis. Healthy controls were attending an ophthalmology clinic for routine eye care and were known to have no current or prior evidence for uveitis. Since medications can markedly affect gene expression, all blood and solid tissue samples were obtained from patients who were not receiving oral corticosteroids or immunomodulatory therapy.
The portion of the study involving peripheral blood included 12 patients with sarcoidosis, 7 of whom also had active uveitis and 12 healthy controls. Healthy individuals were attending an ophthalmology clinic and were thus known to have no active ocular or systemic inflammatory disease. The average age for patients with sarcoidosis was 53.9 ± 12.2 years at the time of enrollment and 48.3 ± 10.9 years at diagnosis. The controls had a mean age of 48.8 ± 21.4 years, which was not statistically different from the patients. Details on gender, race, and methodology for diagnosis are shown in Table 1.
Table 1.
General characteristics of study subjects
| Sarcoidosis diagnosis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject | Age at diagnosis of sarcoidosis | Age at study | Gender | Race | History of uveitis | Chest x-ray* | Chest CT* | Biopsy* | Systemic symptoms |
| Sarcoidosis group | |||||||||
| S1 | 46.9 | 47.5 | F | Caucasian | Yes | N/A | + | N/A | Pulmonary |
| S2 | 49.3 | 49.3 | M | Asian | Yes | + | N/A | None | |
| S3 | 51.7 | 69.6 | F | Caucasian | Yes | N/A | + | N/A | Joint |
| S4 | 39.3 | 41.1 | F | Caucasian | Yes | + | N/A | None | |
| S5 | 64.2 | 68.2 | F | Caucasian | Yes | + | + | + | Pulmonary |
| S6 | 37.6 | 59.1 | F | Caucasian | Yes | + | N/A | N/A | Pulmonary |
| S7 | 25.2 | 27.9 | M | African American | Yes | + | None | ||
| S8 | 47.0 | 58.9 | F | Caucasian | No | N/A | N/A | + | Pulmonary/CNS |
| S9 | 58.6 | 63.2 | F | Caucasian | No | + | + | + | Pulmonary |
| S10 | 44.1 | 44.6 | M | Caucasian | No | + | + | Pulmonary | |
| S11 | 54.9 | 56.9 | F | Caucasian | No | N/A | + | + | Pulmonary |
| S12 | 60.3 | 60.4 | F | Caucasian | No | + | + | N/A | None |
|
| |||||||||
| Control group | |||||||||
| C1 | - | 41.5 | M | Caucasian | No | ||||
| C2 | - | 35.7 | F | Caucasian | No | ||||
| C3 | - | 68.2 | F | Caucasian | No | ||||
| C4 | - | 31.9 | M | Caucasian | No | ||||
| C5 | - | 25.3 | F | Caucasian | No | ||||
| C6 | - | 83.1 | F | Caucasian | No | ||||
| C7 | - | 59.5 | F | Caucasian | No | ||||
| C8 | - | 21.8 | F | Caucasian | No | ||||
| C9 | - | 22.9 | F | Asian | No | ||||
| C10 | - | 69.6 | M | Caucasian | No | ||||
| C11 | - | 68.6 | F | Caucasian | No | ||||
| C12 | - | 57.0 | M | Caucasian | No | ||||
Patients who provided diseased lung or lymph node met the operational diagnosis of sarcoidosis based upon the accepted pathological criterion, i.e., samples displayed well-formed non-necrotizing epithelioid granuloma in the absence of identifiable infection or foreign body, in accordance with diagnostic criteria described in the American Thoracic Society’s joint statement on sarcoidosis [13]. Samples exhibiting atypical pathological features, such as necrosis or fibrosis, were excluded. Disease-free lung tissues were obtained during surgical lung resections, bronchoscopic lung biopsy or in the immediate post-mortem period from patients who had submitted for organ donation for medical research. Each control sample had normal lung histology verified by a certified pathologist. The lymph node samples were from organ donors (normal) or patients undergoing surgical biopsies (sarcoidosis) provided from the Midwestern Division of the Cooperative Human Tissue Network.
Gene expression analysis
Blood was collected directly into PAXGene tubes (2.5 ml/tube; 4 tubes/subject), incubated for at least 2 hours at room temperature for cell lysis and RNA stabilization, and stored at −80 °C. RNA was purified with PAXGene columns and DNase treatment per the manufacturer’s protocol and stored at −80 °C until needed for the microarray procedure. Initial tests confirmed the manufacturer’s claim that there is negligible difference in the microarray hybridization results between samples processed immediately and those frozen in the PAXGene tubes [14].
For studies on peripheral blood, RNA samples were quantified by spectrophotometry using the SpectraMax M2 plate reader (Molecular Devices) and RNA quality was determined using Lab-on-a-Chip RNA NanoChips and the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). This is a capillary electrophoresis system to characterize size distribution. Total RNA quality was verified by the presence of two discrete electropherogram peaks corresponding to the 18S and 28S rRNA at a ratio approaching 2:1. Samples with electropherogram patterns consistent with acceptable microarray performance were selected for labeling and microarray analysis. Three μg of each total RNA were amplified and labeled using the GeneChip Globin Reduction Protocol rev. 1 (Affymetrix, Inc., PreAnalytiX). This protocol uses peptide nucleic acid (PNA) oligonucleotides complementary to human globin mRNA transcripts during the first strand cDNA synthesis reaction of the Affymetrix one-cycle cDNA synthesis/IVT amplification and labeling procedure to improve assay sensitivity by reducing the amount of cDNA generated from globin mRNA. The GeneChip Globin Reduction Protocol was selected after initial studies comparing it with GLOBINclear (Ambion, Austin, TX) and Ovation (NuGEN, SanCarlos, CA) options for minimizing adverse effects from the high globin mRNA content of whole blood (manuscript in preparation). Following cRNA amplification, 10 μg of each labeled target were hybridized with a Human Genome U133 Plus 2.0 array (Affymetrix, Inc.) using standard protocols as described in the GeneChip Expression Analysis manual (www.affymetrix.com/support/technical/manual/expression_manual.affx). The U133 Plus 2.0 array contains 54,000 probe sets designed to analyze the expression of 47,000 human transcripts and variants. Following hybridization, arrays were processed, stained, and then scanned using the GeneChip scanner 3000 (Affymetrix, Inc.). Image processing was performed with the Affymetrix GCOS version 1.4. Initial analysis of individual array performance was performed using the MAS 5.0 statistical analysis program.
Each array scan was processed using the GeneChip Operating Software (GCOS) to produce cell fluorescence intensity (.CEL) files. CEL files were imported into the R statistical language environment [15]. Perfect match (PM) probe data were corrected for background noise using the GeneChip robust multi-array analysis (GCRMA) developed by Wu and co-workers [16]. Corrected PM probe data were normalized with the algorithm based on rank invariant probes by Li and Wong [17]. Gene expression values were determined using a linear model estimated by the median polish algorithm, according to the description of Irizarry and colleagues [18].
After normalization, data sets were compared with the Significance Analysis of Microarrays (SAM) software[19]. This method of analysis is designed for relatively small data sets. It incorporates the concepts of the false discovery rate (FDR) [20] and the q value [21]. The FDR controls for the expected ratio of false positives among significantly expressed genes. The q value is a posterior Bayesian p value, and it indicates the minimum FDR at which the test detects a statistically significant difference. For pair-wise comparisons in this analysis, the FDR was set at 5 %, with a significant difference in gene expression defined as one having a q value less than 0.05. Differentially expressed genes are presented by using heatmaps. All computations were done with R and its add-on packages; “affy”, “gcrma” and “samr” that run above the R environment. The list of genes regulated by the STAT1 transcription factor was extracted from the Transfac Pro Database [22]. The blood data have also been used to illustrate an analytical approach described in a statistical methods paper [23].
Variations in Techniques for Solid Tissue Studies
Frozen tissue was maintained at −80°C until the day before total RNA isolation, at which time the sample was soaked overnight in RNAlater-ICE(Ambion, Applied Biosystems, Foster City, CA) at −20°C. The samples were then removed from RNAlater-ICE and total RNA was isolated using TRIzol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer’s protocol. The RNA was cleaned using the QIAGEN RNeasy Mini Kit (Qiagen Inc., Valencia, CA). The integrity of total RNA samples was assessed qualitatively on an Agilent 2100 Bioanalyzer as above. Following array hybridization, housekeeping genes, β-actin and GAPDH, were used to assess the quality of the synthesized, labeled cRNA. Samples were excluded from gene array analysis if the ratio between the 3′ and 5′ signals exceeded 4, with ideal values being between 1 and 2. Statistical analysis was done by using modified t-test with random variance model using BRB Array Tools software [24]. Q values and false discovery rate adjusted p values are equivalent but differ slightly as the q value uses Bayesian analysis. Studies on blood samples relied on q values and p values were calculated for solid tissue studies because the two centers used different software for normalization and statistical analysis. Additional experimental details and data from the lymph node and lung analysis are in press [25].
Immunohistochemistry
Formalin-fixed, paraffin-embedded lymph nodes were obtained from Oregon Health & Science University pathology archives. Phosphorylated STAT1 (pSTAT1) and non-phosphorylated STAT1 expression were determined by immunohistochemistry on 5 μm sections with purified rabbit polyclonal antibodies detecting either human STAT1 or pSTAT1 at the phosphorylation site of tyrosine 701 (GenScript Corp., Piscataway, NJ, USA). Antigen retrieval was achieved by boiling sections in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 9.0) for 10 minutes. Sections were incubated for 1 hour in blocking solution (4.5% goat serum, 0.36% Triton X-100, 0.1% bovine serum albumin) and then overnight at 4 °C with primary antibodies or control rabbit IgG diluted 1:80 in blocking solution. After several washes, sections were incubated with pre-absorbed, alkaline phosphatase-conjugated anti-rabbit IgG antibody for 1 hour (1:200, GeneTex Inc., San Antonio, TX, USA). Immunostaining was visualized by incubating with Fast Red developer (BioGenex, San Ramon, CA, USA) until coloration was apparent (approx. 2 min.). Sections were counterstained with hematoxylin.
RESULTS
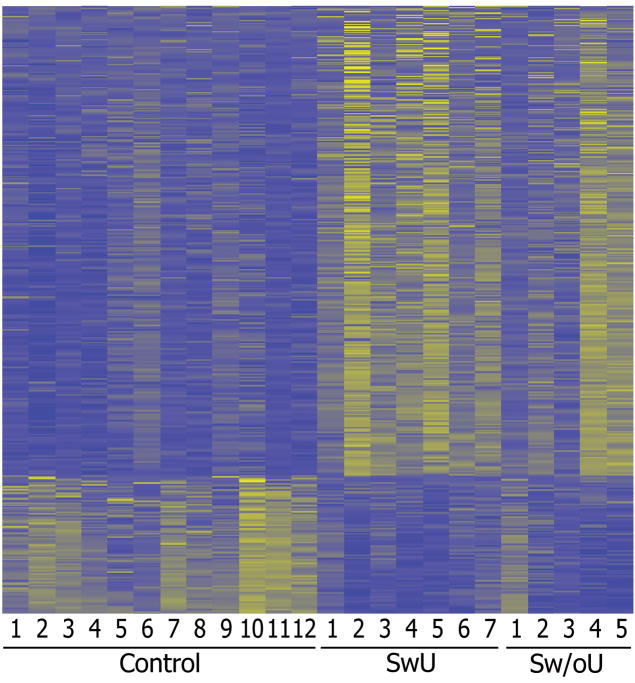
Microarray analysis detected 1187 probe sets (1039 transcripts; many genes in the U133 Plus 2.0 array are represented more than once with different probe sets detecting different portions of the transcript) among the patients with sarcoidosis that were significantly upregulated with q-value less than 0.05 and by a factor of at least 1.5 fold relative to the controls. Twelve hundred eighty-one probes sets (872 transcripts) were downregulated in the sarcoidosis group relative to the controls using the same criteria. The lists of the significantly up- and downregulated genes with at least a 1.5-fold change are shown in Supplementary Tables 1 and 2. The difference between patients with sarcoidosis and controls is shown visually for the probe sets with a minimum of a 2-fold change using a display approach called a heat map which is used in most gene expression studies (Figure 1).
Figure 1.
Heat map differentially expressed transcripts between control and sarcoidosis patients (fold difference ≥ 2 and q ≤ 0.05).436 up- and 128 down-regulated in sarcoidosis groups. Refer to Supplementary Tables 1 and 2 for the complete gene lists.
SwU indicates sarcoidosis with uveitis and Sw/oU indicates sarcoidosis without uveitis.
Our attention focused initially on STAT1 because: 1) the upregulation of the STAT1 transcript could be validated by seven different probe sets; 2) all q values for these probe sets were < 0.002; 3) the average fold change for all STAT1 probe sets was 1.99 with most probe sets showing more than a two-fold increase; and 4) STAT1 is known to be a critical transcription factor in the inflammatory response.
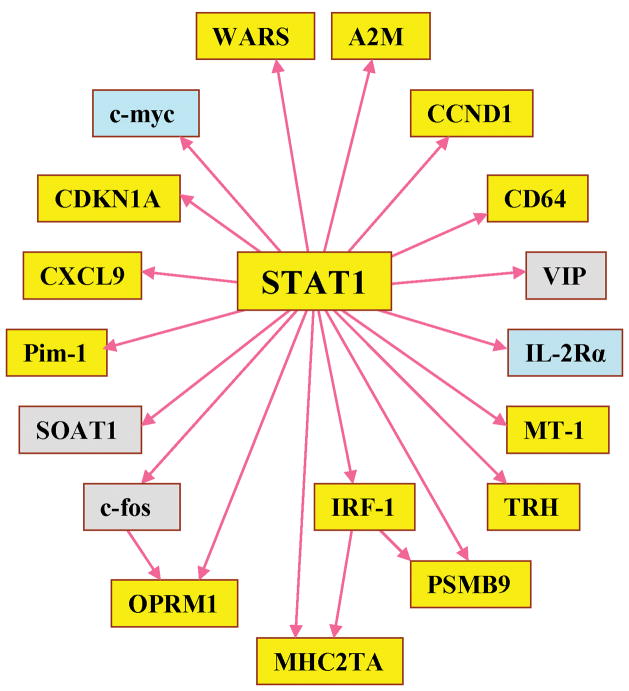
Consequently, we searched the TRANSFAC database to identify genes directly regulated by STAT1. As shown in Figure 2, thirteen of the 18 genes under the regulation of STAT1 were upregulated based on a q value less than 0.05 (Table 2).
Figure 2.
STAT1 downstream genes: yellow genes were significantly upregulated (q-value < 0.05), light blue genes were downregulated, and gray genes were not differentially expressed in the sarcoidosis group compared to the control group.
Table 2.
STAT1-regulated genes with increased expression in blood from patients with sarcoidosis.
| Gene. Symbol | Gene Title | Mean FC* | q-value** | Representative functions |
|---|---|---|---|---|
| MHC2TA | Major histocompatibility complex, class II, transactivator | 1.23 | 0.029 | Master of regulator of MHC class II genes [51] |
| IRF1 | Interferon regulatory factor 1 | 1.45 | 0.002 | Nuclear factor and a transcriptional factor for type I IFN [52], can induce CDKN1A activity |
| WARS | Tryptophanyl-tRNA synthetase | 1.72 | 0.001 | Antagonist of VEGF [53] |
| PSMB9 | Proteasome subunit, beta-type, 9 | 1.42 | 0.002 | Transport protein into the endoplasmic reticulum, important in MHC class I regulation [54] |
| CD64 | Fc fragment of IgG, high affinity | 3.14 | 0.029 | Fc fragment of IgG receptor, expressed on human monocytes and macrophages [55] |
| CXCL9 | Chemokine CXC motif, ligand 9 | 1.16 | 0.029 | T-cell chemoattractant |
| PIM1 | Oncogene PIM1 | 1.39 | 0.009 | Protooncogene in prostate cancer and hematopoietic malignancies [56] |
| CCND1 | Cyclin D1 | 1.16 | 0.029 | Function as an oncogene, related to lymphoma, leukemia [57], parathyroid tumor [58], and colorectal cancer [59] |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | 1.36 | 0.029 | Inhibitor of protein kinase, CDK1 |
| A2M | Alpha-2-macroglobulin | 1.15 | 0.029 | Major plasma proteinase inhibitor |
| OPRM-1 | Opioid receptor, mu-1 | 1.15 | 0.029 | Pain-related gene |
| MT1E | Metallothionein 1E | 1.31 | 0.012 | Heavy metal-binding protein, Zn and Cu homeostasis |
| TRH | Thyrotropin-releasing hormone deficiency | 1.14 | 0.029 | Major hypothalamic mediator of thyroid stimulating hormone release |
Fold change,
the maximum q-value in case of multiple probe sets
We additionally analyzed other STATs, JAKs (the kinase known to activate STATs), and interferon receptors and noted that many were consistently upregulated in the patients with sarcoidosis relative to controls (Table 3). The interferon regulatory factors (IRFs) are a family of 9 transcription factors which are activated by interferons and other inflammatory mediators. Eight of the 9 known IRFs are detected by the microarray employed in our study. Transcripts for six of the 8 detectable IRFs (IRF 1, 2, 4, 5, 6, and 7) were upregulated (range 1.13 to 2.39 fold) with a q value <0.05 for each in the peripheral blood of sarcoidosis patients compared to controls.
Table 3.
Genes in JAK-STAT pathway upregulated in the peripheral blood of patients with sarcoidosis.
| Gene symbols | Gene titles | Number of probe sets | Mean fold increase | q-value* |
|---|---|---|---|---|
| IFNAR1 | interferon (alpha, beta and omega) receptor 1 | 3 | 1.25 | 0.029 |
| IFNAR2 | interferon (alpha, beta and omega) receptor 2 | 2 | 1.28 | 0.028 |
| IFNGR2 | interferon γreceptor 2 (interferon γ transducer 1) | 1 | 1.11 | 0.029 |
|
| ||||
| JAK1 | Janus kinase 1 (a protein tyrosine kinase) | 3 | 0.88 | 0.039 |
| JAK2 | Janus kinase 2 (a protein tyrosine kinase) | 3 | 1.38 | 0.029 |
|
| ||||
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 6 | 1.99 | 0.001 |
| STAT2 | signal transducer and activator of transcription 2, 113kDa | 3 | 1.44 | 0.029 |
| STAT3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 4 | 1.38 | 0.029 |
Number of Probe Sets reflects the characteristics of the Affymetrix U133 Plus 2.0 array which detects some transcripts with discrete probes that identify different portions of the transcript.
The maximum q-value in case of multiple probe sets
We included two additional checks on the validity of our analysis. We studied a group of 8 patients with idiopathic uveitis. All patients in this group had active intraocular inflammation, but they also presumably represent inflammation that has resulted from a variety of different etiologies. In this group, we found only 6 statistically significant differences in gene expression between patients and the control group. The upregulation of 6 transcripts is possibly not pathogenetically significant, as one would expect to find by chance at least 6 differences between data sets based solely on the number of statistical comparisons performed. We also studied patients with ankylosing spondylitis (n = 12; 7 with uveitis and 5 without uveitis). While these patients had a gene expression pattern that differs from controls, it did not reflect a pattern of genes regulated by STAT1 (manuscript in preparation).
Studies based on relatively small numbers of subjects and involving multiple statistical comparisons are fraught with the detection of differences that are not reproducible (i.e., type 1 statistical errors). The finding that many transcripts regulated by STAT1 are also increased strongly suggests that increased STAT1 mRNA is not an artifact. However, as an additional validation, we compared the STAT1 signature in peripheral blood with gene expression in two tissues classically affected in sarcoidosis, lung and lymph node. Demographics for these patients are shown in Table 4. In addition to upregulation of STAT1 itself, six of the 18 STAT1 regulated genes as identified by the TRANSFAC database showed increased expression in lymph node based on FDR adjusted p<0.05 for each comparison. The upregulated transcripts were CXCL9, IRF-1, A2M, WARS, c-fos, and SOAT1. The transcript for the chemokine, CXCL9 was upregulated 9 fold. This same transcript was upregulated 9.5 fold in the lung. The TRANSFAC database used to analyze the data from peripheral blood does not include CXCL10 and CXCL11 in its listing of STAT1 regulated genes, although CXCL10 is γ-interferon inducible protein 10 (IP10) and CXCL11 is γ-interferon inducible T cell alpha chemoattractant (I-Tac). Table 5 summarizes the upregulation of STAT1 and the transcripts for CXCL9, CXCL10, and CXCL11 as found in blood, lymph node, and lung relative to the appropriate control tissue.
Table 4.
Patient Demographics for Tissue Gene Expression Analyses
| Lung | Lymph node | |||
|---|---|---|---|---|
| Control (n = 6) | Sarcoidosis (n = 6) | Control (n=5) | Sarcoidosis (n=8) | |
| Age ± SEM (yrs) | 50.8 ± 5.2 | 40.7 ± 5.6 | 43.0 ± 6.1 | 40.0 ± 4.8 |
| Gender (male/female) | 3/3 | 2/4 | 3/2 | 3/5 |
| Race (White/Black/Other) | 4/2/0 | 4/2/0 | 3/2/0 | 5/3/0 |
Table 5.
Upregulation of Transcripts for STAT1 regulated chemokines in sarcoidosis. Values indicate fold upregulation compared to gene expression in control tissue.
| STAT1 | CXCL9 | CXCL10 | CXCL11 | |
|---|---|---|---|---|
| Blood | 2.0*** | 1.2* | 2.8** | 1.1* |
| Lung | 4.3*** | 9.5*** | 5.1** | 9.8*** |
| Lymph node | 7.0*** | 19.5** | NS | 19.2** |
p or q <0.03,
p or q <0.01,
p or q <0.001 (q values for blood samples and the FDR adjusted p values for lung and lymph node samples.) NS=not significant
Although STAT2 and STAT3 were slightly upregulated in peripheral blood, we were unable to show that these two specific transcription factors or other STATs were up regulated in the lymph nodes from patients with sarcoidosis.
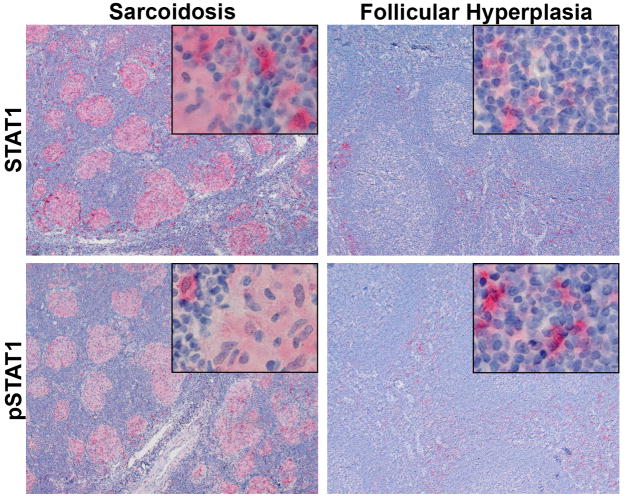
All of the above studies are based on the measurement of mRNA. In order to determine if the alteration in mRNA expression correlated with a change in protein expression, we examined the presence of both STAT1 and activated STAT1 (phosphorylated STAT1) in lymph nodes from 6 patients with sarcoidosis and 6 control subjects with follicular hyperplasia. Both STAT1 and pSTAT1 were much more abundant in the lymph nodes of patients with sarcoidosis than in control lymph nodes (Figure 3). In addition, the expression of STAT1 was predominantly in the granulomas themselves suggesting that it is directly involved in the pathogenesis of the granulomas.
Figure 3.
Sarcoidosis patients have abundant STAT1 and pSTAT1 in lymph node granulomas. Sections of lymph nodes from patients with sarcoidosis or follicular hyperplasia (controls) were immunostained for STAT1 or pSTAT1. Both sarcoid and control lymph nodes had scattered cortical STAT1 and pSTAT1 positive cells. In addition, granulomas in the sarcoid samples were strongly positive for both forms of STAT1. No staining was evident when control IgG was substituted for the primary antibodies (not shown). Original magnification main panels 100X; inserts 400X.
DISCUSSION
Our results indicate that RNA for the major transcription factor, STAT1, is upregulated in the peripheral blood of patients with sarcoidosis compared to healthy controls. In addition, mRNAs for 13 of the 18 genes directly regulated by STAT1 have a statistically significant increase in the blood of patients with sarcoidosis. There are 7 known STATs which join in various combinations of homo- or heterodimers [10]. The STATs are activated by Janus protein tyrosine kinases (JAKs). Although signaling through many receptors is dependent on JAK-STAT activation, interferons are especially dependent on this pathway. γ-interferon binds to a specific receptor and induces gene expression via activation of STAT1 homodimers.
A limitation of our study is that the sample size is relatively small and our analysis involves multiple statistical comparisons. However, the upregulation of this transcript is confirmed by multiple independent probe sets which are included in the microarray. All but one of these probes indicated that the transcript for STAT1 was significantly upregulated. More importantly the proposed role of STAT1 is strongly supported by RNA data from lung and lymph node and the detection of activated STAT1 protein in granulomas from lymph nodes of patients with sarcoidosis. Although STAT1 has not been implicated previously in the pathogenesis of sarcoidosis, it may be a major contributor to many clinical characteristics. First, the increase in STAT1 fits well with a possible infectious trigger such as mycobacteria for this disease.
Catalase-peroxidase derived from M. tuberculosis was found in affected tissue of 55% of patients with sarcoidosis and in no control tissue [26]. Mycobacterial antigens strongly induce the production of γ-interferon [27]. Polymorphisms in the STAT1 gene influence susceptibility to mycobacterial infection [28]. Mice which lack STAT1 are especially susceptible to mycobacterial infection in the lung [29]. Mice infected with the parasite, Angiostrongylus cantonensis, have increased STAT1 in their granulomatous brains [30]. Second, activated expression of STAT1 explains why sarcoidosis is generally considered to be a TH1-mediated disease. T cell subsets are grouped on the basis of the most abundant cytokines which are produced. T cells which best express γ-interferon are designated TH1. This subset of T cells predominates in the broncho-alveolar lavage of patients with sarcoidosis [31; 32; 33]. Since γ-interferon induces STAT1 expression, a TH1 mediated disease should be associated with increased expression of STAT1. On the other hand, TH1 expressing lymphocytes do not predominate in the peripheral blood of patients with sarcoidosis [32]. Accordingly our peripheral blood measurements likely reflect sources in addition to T cells. Third, hypercalcemia, a well described complication of sarcoidosis, can be explained by an increased expression of STAT1 [34], which enhances the conversion of 25-hydroxy vitamin D to its more active form, 1, 25 di-hydroxyvitamin D. Fourth, independent studies have already noted an increase in protein in the serum of patients with sarcoidosis for several chemokines regulated by STAT1 including CXCL9 (MIG or monokine induced by γ-interferon) and CXCL10 (IP10, interferon inducible protein 10) [35]. Fifth a published study that used microarray to study bronchoalveolar lavage cells from 3 patients with sarcoidosis found elevation of TYK2 and p21Waf1/Cip1 [36]. Both of which are regulated by γ-interferon and thus are consistent with our observations. Finally, more than 60 patients have developed granulomatous disease subsequent to treatment with various interferons [37; 38; 39]. This clinical condition mimics sarcoidosis.
The potential importance of STAT1 in the pathogenesis of sarcoidosis is supported by observations in both lymph node and lung. However, the findings in solid tissue are not identical to those from blood, i.e., not all STAT1 regulated transcripts showing upregulation in blood showed statistically significant upregulation in solid tissue. Since cellular composition and local factors within specific organs or tissues will modify gene expression, these differences are not surprising. Gene expression in blood resembles gene expression in lymph node more than lung, consistent with greater cell trafficking between blood and lymph node. Tissue response to a cytokine such as interferon obviously depends on cells which are present in that tissue. For example, even adjacent cells such as astrocytes and microglia differ in their synthesis of CXCL9 and CXCL10 [40]. Nonetheless, STAT1 regulated genes are strongly represented in all the sarcoidosis tissues that we examined.
CXCL9 was consistently upregulated in blood, lymph node, and lung. Like CXCL10, CXCL9 binds to a receptor known as CXCR3. CXCL9 has been strongly implicated in granuloma formation in primates [41]. CXCL9 plays a major role in several inflammatory diseases including autoimmune diseases of the skin [42; 43], inflammatory bowel disease [44], forms of arthritis [45], demyelinating disease [46], and several infections [47; 48].
As indicated in Supplementary Table 1, STAT1 is far from the only gene upregulated in the blood of patients with sarcoidosis. For example, we found upregulation of the receptor for epidermal growth factor, which has been associated with mycobacterial-induced granulomas [49]. Our analysis does not negate the importance of other genes or gene networks in the pathogenesis of this disease. Rather, our findings clearly support multiple factors in the pathogenesis.
Sarcoidosis is not the first immune mediated disease for which interferon regulated genes have been implicated. Gene expression studies have also suggested a role for genes regulated by either interferon type 1or γ-interferon in immunological diseases which include rheumatoid arthritis, inflammatory myopathies, and SLE [6; 9]. STAT1 is upregulated in the affected skin of patients with psoriasis [50] and in the synovium of patients with rheumatoid arthritis [51]. Polymorphisms in STATs have also been linked to susceptibility of several immune-mediated diseases including Graves’ disease, SLE, and rheumatoid arthritis [52; 53]. The role of STAT1 polymorphisms in the susceptibility to sarcoidosis has not been thoroughly studied.
The pattern of gene expression could conceivably have diagnostic, prognostic, and therapeutic implications. Further study will determine how therapy impacts gene expression and if gene expression predicts response to a specific intervention. Additional study is indicated to determine which cells are most responsible for the detection of mRNA for STAT1 in our studies. Certainly if a role for STAT1 is confirmed, medications that target STAT1 could be considered to treat sarcoidosis.
Supplementary Material
Acknowledgments
We are indebted to Kristina Vartanian and Rachel Slottke for technical support and to Cathy Markin for identification of patients with active pulmonary sarcoidosis.
Support: NIH Grants, HL077466, EY015858, EY010572; Research to Prevent Blindness Awards to the Casey Eye Institute and to JTR, SRP, and JRS; and the Stan and Madelle Rosenfeld Family Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Culver DA, Newman LS, Kavuru MS. Gene-environment interactions in sarcoidosis: challenge and opportunity. Clin Dermatol. 2007;25:267–275. doi: 10.1016/j.clindermatol.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spagnolo P, DuBois RM. Genetics of sarcoidosis. Clin Dermatol. 2007;25:242–249. doi: 10.1016/j.clindermatol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 4.Dai DL, Wang Y, Liu M, Martinka M, Li G. Bim expression is reduced in human cutaneous melanomas. Invest Dermatol. 2008;128:403–7. doi: 10.1038/sj.jid.5700989. [DOI] [PubMed] [Google Scholar]
- 5.Driouch K, Landemaine T, Sin S, Wang S, Lidereau R. Gene arrays for diagnosis, prognosis, and treatment of breast cancer metastasis. Clin Exp Metastasis. 2007;24:575. doi: 10.1007/s10585-007-9110-x. [DOI] [PubMed] [Google Scholar]
- 6.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szodoray P, Alex P, Frank MB, Turner M, Turner S, Knowlton N, Cadwell C, Dozomorov I, Tang Y, Wilson PC, Jonsson R, Centola M. A genome-scale assessment of peripheral blood B-cell molecular homeostasis in patients with rheumatoid arthritis. Rheumatology. 2006;45:1466–1476. doi: 10.1093/rheumatology/kel095. [DOI] [PubMed] [Google Scholar]
- 8.Mandel M, Gurevich M, Pauzner R, Kaminski N, Achiron A. Autoimmunity gene expression portrait: specific signature that intersects or differentiates between multiple sclerosis and systemic lupus erythematosus. Clin Exp Immunol. 2004;138:164–170. doi: 10.1111/j.1365-2249.2004.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, Ytterbert SR, Gregersen PK, Behrens TW, Reed AM. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007;13:59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brierly MM, Fish EN. Stats: Multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- 11.Sweiss NJ, Curran J, Baughman RP. Sarcoidosis, role of tumor necrosis factor inhibitors and other biologic agents, past, present, and future concepts. Clin Dermatol. 2007;25:341–346. doi: 10.1016/j.clindermatol.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Winterbauer RH, Belic N, Moores KD. Clinical interpretation of bilateral hilar adenopathy. Ann Intern med. 1973;78:65–71. doi: 10.7326/0003-4819-78-1-65. [DOI] [PubMed] [Google Scholar]
- 13.Society AT. American Thoracic Society Statement on Sarcoidosis. Am J Respir Crit Care Med. 1999;160:736–755. doi: 10.1164/ajrccm.160.2.ats4-99. [DOI] [PubMed] [Google Scholar]
- 14.Vartanian K, Slottke R, Johnstone T, Casale A, Planck SR, Choi D, Smith JR, Rosenbaum JT, Harrington CA. Gene expression profiling of whole blood: Comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10:2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Team RDC. R: a language and environment for statistical computing R Foundation for Statistical Computing. Viena. 2007 [Google Scholar]
- 16.Wu Z, Irizarry RA, Gentleman R, et al. A model-based background for adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 17.Li W, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochbert Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 21.Storey J. A direct approach to false discovery rates. J R Stat Soc, B. 2002;64:479–498. [Google Scholar]
- 22.Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi D, Sharma SM, Pasadhika S, Kang Z, Harrington CA, Smith JR, Planck SR, Rosenbaum JT. Application of Biostatistics and Bioinformatics Tools to Identify Putative Transcription Factor-Gene Regulatory Network of Ankylosing Spondylitis and Sarcoidosis. Communications in Statistics - Theory and Methods. 2009 doi: 10.1080/03610920902898472. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 25.Crouser ED, Culver DA, Knox KS, Julian MW, Shao G, Abraham S, Liyanarachchi S, Marcre JE, Wewers MD, Gavrilin MA, Ross P, Abbas A, Eng C. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2009 doi: 10.1164/rccm.200803-490OC. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z, Marzilli L, Greenlee BM, Chen ES, Silver RF, Askin FB, Teirstein AS, Zhang Y, Cotter RJ, Moller DR. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J Exp Med. 2005;201:755–767. doi: 10.1084/jem.20040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlisle J, Evans W, Hajizadeh R, Nadef M, Shepherd B, Ott RD, Richter K, Drake W. Multiple mycobacterium antigens induce interferon-gamma production from sarcoidosis peripheral blood mononuclear cells. Clin Exp Immunol. 2007 doi: 10.1111/j.1365-2249.2007.03510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, Santos OF, Bustamante J, Picard C, DeBeaucoudrey L, Emile JF, Arkwright PD, Schreiber RD, Rolinck-Werninghaus C, Rosen-Wolff A, Magdorf K, Roesler J, Casanova JL. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugawara I, Yamada H, Mizuno S. STAT1 knockout mice are highly susceptible to pulmonary mycobacterial infection. J Exp Med. 2004;202:41–50. doi: 10.1620/tjem.202.41. [DOI] [PubMed] [Google Scholar]
- 30.Lan KP, Wang CJ, Hsu JD, Chen KM, Lai SC, Lee HH. Induced eosinophilia and proliferation in Angiostrongylus cantonensis-infected mouse brain are associated with the induction of JAK/STAT1, IAP/NF-?B and MEKK1/JNK signals. Journal of Helminthology. 2004;78:311–317. doi: 10.1079/joh2004256. [DOI] [PubMed] [Google Scholar]
- 31.Antoniou KM, Tzouvelekis A, Alexandrakis MG, Tsiligianni I, Tzanakis N, Sfiridaki K, Rachiotis G, Bouros D, Siafakas NM. Upregulation of Th1 cytokine profile (IL-12, IL-18) in bronchoalveolar lavage fluid in patients with pulmonary sarcoidosis. J Interferon Cytokine Res. 2006;26:400–405. doi: 10.1089/jir.2006.26.400. [DOI] [PubMed] [Google Scholar]
- 32.Inui N, Chida K, Suda T, Nakamura H. TH1/TH2 and TC1/TC2 profiles in peripheral blood and bronchoalveolar lavage fluid cells in pulmonary sarcoidosis. J Allergy Clin Immunol. 2001;107:337–344. doi: 10.1067/mai.2001.112273. [DOI] [PubMed] [Google Scholar]
- 33.Prasse A, Georges CG, Biller H, Hamm H, Matthys H, Luttmann W, Virchow JCJ. Th1 cytokine pattern in sarcoidosis is expressed by bronchoalveolar CD4+ and CD8+ cells. Clin Exp Immunol. 2000;122:241–248. doi: 10.1046/j.1365-2249.2000.01365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overbergh L, Stoffels K, Waer M, Verstuyf A, Bouillon R, Mathieu C. Immune regulation of 25-hydroxyvitamin D-1alpha-hydroxylase in human monocytic THP1 cells: mechanisms of interferon-gamma-mediated induction. J Clin Endocrinol Metab. 2006;91:3566–3574. doi: 10.1210/jc.2006-0678. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi M, Oh-I K, Suzuki J, Hattori T, Takeuchi A, Okunuki Y, Usui Y, Usui M. Elevated serum levels of CXCL9/monokine induced by interferon-gamma and CXCL10/interferon-gamma-inducible protein-10 in ocular sarcoidosis. Invest Ophthalmol Vis Sci. 2006;47:1063–1068. doi: 10.1167/iovs.05-0966. [DOI] [PubMed] [Google Scholar]
- 36.Schischmanoff PO, Naccache JM, Carrere A, Richardson S, Kambouchner M, Raphael M, Valeyre D, Fagard R. Progressive pulmonary sarcoidosis is associated with over-expression of TYK2 and p21Waf1/Cip1. Sarcoidosis Vasc Diffuse Lung Dis. 2006;23:101–107. [PubMed] [Google Scholar]
- 37.Alazemi S, Campos MA. Interferon-induced sarcoidosis. Int J Clin Pract. 2006;60:201–211. doi: 10.1111/j.1742-1241.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 38.Fiel MI, Shukla D, Saraf N, Xu R, Shciano TD. Development of hepatic granulomas in patients receiving pegylated interferon therapy for recurrent hepatitis C virus post liver transplantation. Transpl Infect Dis. 2008;10:184–9. doi: 10.1111/j.1399-3062.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg HJ, Fiedler D, Webb A, Jagirdar J, Hoyumpa AM, Peters J. Sarcoidosis after treatment with interferon-alpha: a case series and review of the literature. Respir Med. 2006;100:2063–2068. doi: 10.1016/j.rmed.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Carter SL, Muller M, Manders PM, Campbell IL. Induction of the genes for Cxcl9 and Cxcl10 is dependent on IFN-gamma but shows differential cellular expression in experimental autoimmune encephalomyelitis and by astrocytes and microglia in vitro. Glia. 2007;55:1728–1739. doi: 10.1002/glia.20587. [DOI] [PubMed] [Google Scholar]
- 41.Fuller CL, Flynn JL, Reinhart TA. In situ study of abundant expression of proinflammatory chemokines and cytokines in pulmonary granulomas that develop in cynomolgus macaques experimentally infected with Mycobacterium tuberculosis. Infection and Immunity. 2003;71:7023–7034. doi: 10.1128/IAI.71.12.7023-7034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mee JB, Johnson CM, Morar N, Burslem F, Groves RW. The psoriatic transcriptome closely resembles that induced by interleukin-1 in cultured keratinocytes: dominance of innate immune responses in psoriasis. Am J Path. 2007;171:32–42. doi: 10.2353/ajpath.2007.061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wenzel J, Peters B, Zahn S, Birth M, Hofmann K, Kusters D, Tomiuk S, Baron JM, Merk HF, Mauch C, Krieg T, Bieber T, Tuting T, Bosio A. Gene expression profiling of lichen planus reflects CXCL9+-mediated inflammation and distinguishes this disease from atopic dermatitis and psoriasis. J Invest Dermatol. 2008;128:67–78. doi: 10.1038/sj.jid.5700945. [DOI] [PubMed] [Google Scholar]
- 44.Egesten A, Eliasson M, Olin AI, Erjefait JS, Bjartell A, Sangfelt P, Carlson M. The proinflammatory CXC-chemokines GRO-alpha/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J Colorectal Dis. 2007;22:1421–1427. doi: 10.1007/s00384-007-0370-3. [DOI] [PubMed] [Google Scholar]
- 45.Shin JJ, Glickstein LJ, Steere AC. High levels of inflammatory chemokines and cytokines in joint fluid and synovial tissue throughout the course of antibiotic-refractory lyme arthritis. Arthritis Rheum. 2007;56:1325–1335. doi: 10.1002/art.22441. [DOI] [PubMed] [Google Scholar]
- 46.Frisullo G, Angelucci F, Cagguila M, Nociti V, Iorio R, Patanella AK, Sancricca C, Mirabella M, Tonali PA, Batocchi AP. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J Neurosci Res. 2006;84:1027–1036. doi: 10.1002/jnr.20995. [DOI] [PubMed] [Google Scholar]
- 47.Johnson LM, Scott P. STAT1 expression in dendritic cells, but not T cells, is required for immunity to Leishmania major. J Immunol. 2007;178:7259–7266. doi: 10.4049/jimmunol.178.11.7259. [DOI] [PubMed] [Google Scholar]
- 48.Anathbandhu C, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD. STAT1 signaling modulates HIV-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood. 2008;111:2060–2072. doi: 10.1182/blood-2007-05-091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bermudez L, Petrofsky M, Shelton K. Epidermal growth factor-binding protein in mycobacterium avium and mycobacterium tuberculosis: a possible role in the mechanism of infection. Infect Immun. 1996;64:2917–2922. doi: 10.1128/iai.64.8.2917-2922.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Fits L, van der Wei LI, Laman JD, Prens EP, Verschuren MC. In psoriasis lesional skin the type I interferon signaling pathway is activated, whereas interferon-alpha sensitivity is unaltered. J Invest Dermatol. 2004;122:51–60. doi: 10.1046/j.0022-202X.2003.22113.x. [DOI] [PubMed] [Google Scholar]
- 51.Kasperkovitz PV, Verbeet NL, Smeets TJ, van Rietschoten JG, Kraan MC, van der Pouw Kraan TC. Activation of the STAT1 pathway in rheumatoid arthritis. Ann Rheum Dis. 2004;63:233–239. doi: 10.1136/ard.2003.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Remmers EF, Plenge RM, Lee AT, Graham RR, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Land KJ, Moll JS, Kaplan MH, Seetharamaiah GS. Signal transducer and activator of transcription (Stat)-6-dependent, but not Stat4-dependent, immunity is required for the development of autoimmunity in Graves’ hyperthyroidism. Endocrinology. 2004;145:3724–3730. doi: 10.1210/en.2004-0352. [DOI] [PubMed] [Google Scholar]
- 54.Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harada H, et al. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 56.Otani A, et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci USA. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Kaer L, et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 58.Pearse RN, Feinman R, Ravetch JV. Characterization of the promoter of the human gene encoding the high-affinity IgG receptor: transcriptional induction by gamma-interferon is mediated through common DNA response elements. Proc Natl Acad Sci USA. 1991;88:11305–11309. doi: 10.1073/pnas.88.24.11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amson R, et al. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci USA. 1989;86:8857–8861. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsujimoto Y, et al. Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(11;14) chromosome translocation. Science. 1984;224:1403–1406. doi: 10.1126/science.6610211. [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg CL, et al. Rearrangement and overexpression of D11S287E, a candidate oncogene on chromosome 11q13 in benign parathyroid tumors. Oncogene. 1991;6:449–453. [PubMed] [Google Scholar]
- 62.Kong S, et al. Cyclin D1 polymorphism and increased risk of colorectal cancer at young age. J Natl Cancer Inst. 2001;93:1106–1108. doi: 10.1093/jnci/93.14.1106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.