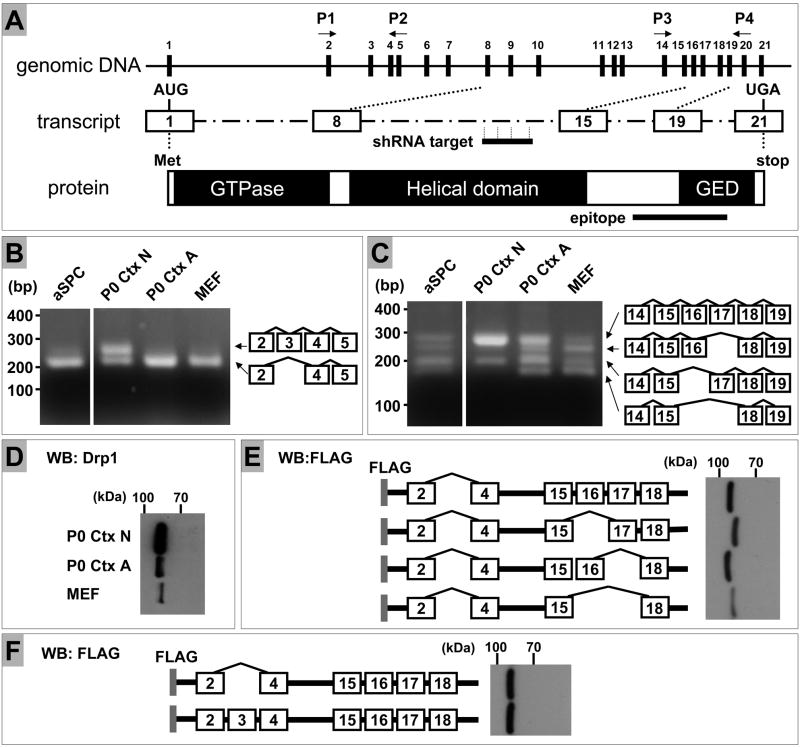
Figure 3. Analysis of expression of Drp1 splicing variants in mouse cells.
(A) Schematic representation of genomic organization, transcript and protein of mouse Drp1. Numbering of exons is based on the sequence of transcript variant “a” (GenBank accession No., NM152816). shRNA target sequence resides in the region corresponding to exon 11, whereas the peptide sequence used to raise the Drp1 antibody we used encompasses the region corresponding to exon 18–20. Thus, all of the splicing variants and the respective proteins can be targeted by our shRNA sequence and recognized by the antibody used in this study, respectively.
(B) Total RNA was prepared from adult spinal cord neural progenitor cells (aSPC), primary neuronal cultures derived from postnatal day 0 cortex (P0 Ctx N), astrocyte cultures from postnatal day 0 cortex (P0 Ctx A) and mouse embryonic fibroblasts (MEF). Each sample was subjected to RT-PCR analysis using primers P1 and P2 for amplification of exon 2–5. The resultant reaction mixtures were resolved on 2.0 % agarose gels with markers (100 bp PCR Molecular Ruler, Bio-Rad. Hercules, CA). The structure of each splicing variant was confirmed by direct sequencing.
(C) Total RNA was prepared and subjected to RT-PCR analysis using primers P3 and P4 for amplification of exon 14–19, followed by agarose gel analysis as described above in (B). The aSPC and other samples were applied to the same agarose gel. The resultant photo images, which contained unrelated samples, were cut and rearranged to facilitate the comparison.
(D) Protein samples prepared as described in Fig. 2A were subjected to SDS polyacrylamide gel electrophoresis using a constant 7.5% gel followed by Western blot analysis. Drp1 protein from MEF migrated faster than those from P0 cortical neurons (P0 Ctx N) and astrocytes (P0 Ctx A).
(E) HEK 293 FT cells were transiently transfected with plasmids harboring FLAG-tagged human Drp1 cDNA with all combinations of inclusion/exclusion of exon 16 and 17. All of these cDNAs lack exon 3. Protein extracts from transfectants were subjected to Western blot using a constant SDS 7.5% polyacrylamide gel to detect FLAG-tagged proteins. The DNA sequences corresponding to exon 16 and 17 encode 26 and 11 amino acids, respectively. Exclusion of exon 16 yielded a faster migrating form as expected, whereas inclusion or exclusion of exon 17 did not provide separable migration patterns under these conditions. Since neuronal Drp1 is expected to be predominantly the form that includes both exon 16 and 17 (C), exclusion of exon 16, not exon 17, can explain the faster moving Drp1 from MEF relative to the neuronal Drp1.
(F) FLAG-tagged human Drp1 cDNAs containing both exon 16 and 17 with/without exon 3 were used in the same way as described in (E). The DNA sequence corresponding to exon 3 encodes 13 amino acids. Inclusion or exclusion of exon 3 did not provide separable migration patterns under these conditions.