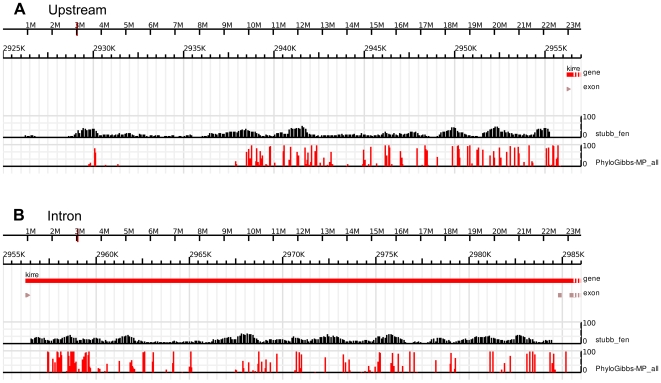
Figure 5. Stubb and PhyloGibbs-MP module predictions for duf genomic region.
Using 31 position weight matrices constructed from the FlyReg database, from B1H data (Noyes et al.), and from other sources, for mesoderm-relevant transcription factors, Stubb predicts a modular structure both in the upstream and in the intronic region. PhyloGibbs-MP, run in module-prediction mode, predicts a similar modular structure. In particular, much of the first 20 kb upstream of duf appears to be potentially regulatory, with significant modular structure in the first 10 kb upstream and again 15–18 kb upstream of the transcriptional start site. Another enhancer appears about 25 kb upstream of the transcriptional start, which is only weakly picked up by PhyloGibbs-MP, suggesting that it may bind a different set of factors. Both programs pick up significant modules at both ends of the intron and some isolated clusters in the middle.