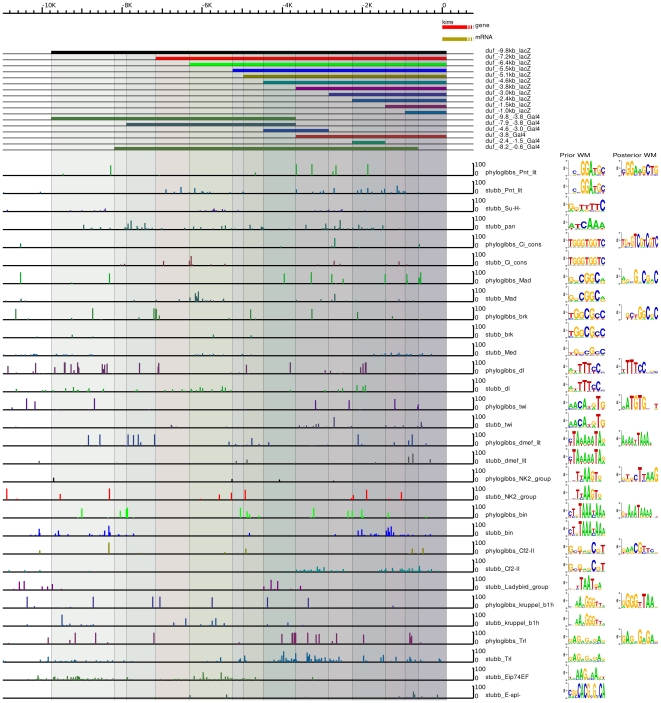
Figure 6. PhyloGibbs-MP and Stubb results for 11 kb duf upstream.
Predictions of individual binding sites, for various factors of interest, from Stubb and PhyloGibbs-MP along with different duf enhancer reporter deletion constructs. Stubb predicts a binding affinity of a known factor using its previously characterised position weight matrix, while PhyloGibbs-MP predicts sites that are similar to one another, but allows its search to be biased via “informative prior” weight matrices. For both programs, position weight matrices for 31 mesoderm-relevant factors, as discussed in the text, were used as priors. These are shown in the left column of sequence logos, marked “Prior WM”. PhyloGibbs-MP in addition reports a posterior WM for all motifs it finds, which is a base-count of all sites reported in each motif, weighted by their significance. Sequence logos for these are in the right column, “Posterior WM”. Stubb reports no posterior WM. PhyloGibbs-MP′s posterior WMs are, in general, similar to the prior WMs, at least in their core features. Moreover, several positions in each PWM have high information scores, indicating a high degree of similarity among the predicted sites. These two facts encourage confidence in the quality of PhyloGibbs-MP′s site predictions as well as in the associations with the prior WMs. Nevertheless, as with any bioinformatics program, some false positives are expected, and several genuine sites may have been missed. The sequence logos were made with Weblogo 2.8 [75]. The predictions were plotted with our genome visualisation tool (S. Acharya and R. Siddharthan, unpublished).