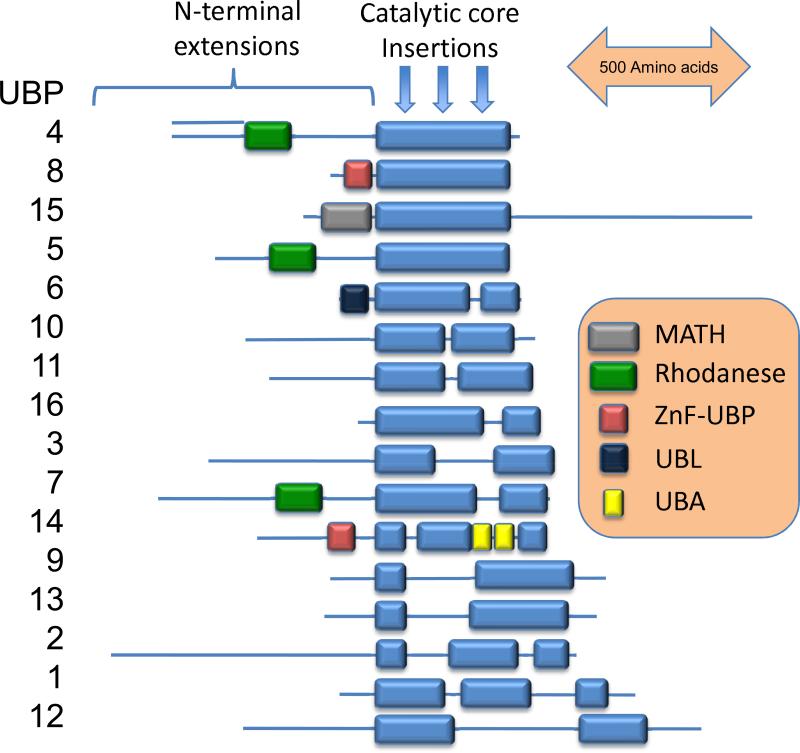
Figure 1.
Domain structure of yeast UBP deubiquitinating enzymes. The catalytic core is indicated by the blue boxes. Other common domains are indicated with differently colored boxes: MATH, meparin and TRAF homology; Rhodanese, rhodanese-like; ZnF-UBP, zinc finger common in UBP DUBs, UBL, ubiquitin-like, and UBA, ubiquitin-associated. Three sites of common insertions within the catalytic core are indicated by arrows. These insertions and the N- and C-terminal extensions are thought to provide specific sequences that define substrate specificity and provide interaction surfaces for binding to adapters and scaffolds.