FIG. 4.—
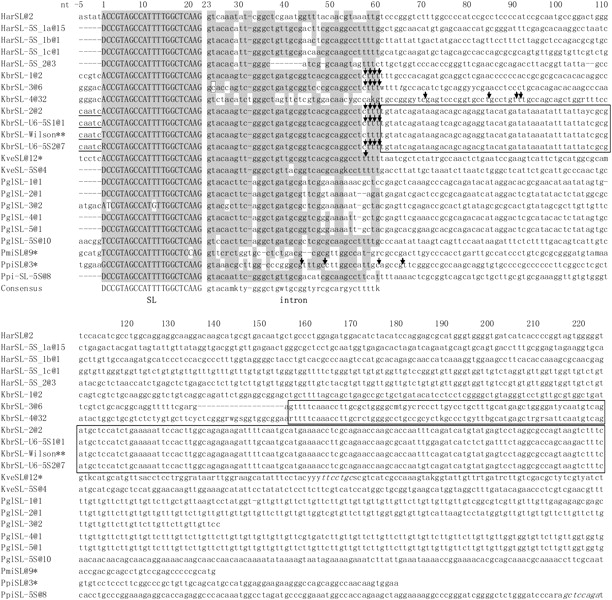
Conservation of the ∼60-bp dinoflagellate SL RNA coding region (SL + intron). In the alignment are representative sequences for each type; the number of identical clones retrieved for each type is indicated by “@number” following the species abbreviation and type number. Har, Heterocapsa arctica; Kbr, Karenia brevis; Kve, Karlodinium veneficum; Ppi, Pfiesteria piscicida; Pgl, Polarella glacialis; and Pmi, Prorocentrum minimum. SL refers to SL RNA sequences obtained from SL-only repeats; SL–5S indicates SL RNA sequences from genes associated with 5S rRNA genes. *: Sequence from Zhang, Hou, et al. (2007); **: sequence from Lidie and van Dolah (2007). SL RNAs mapped by 3′-RACE analyses are denoted by arrows to indicate the terminal positions. The 22-nt SL (exon) was shown in uppercase letters, whereas the intron and the downstream region were in lowercase letters. Shaded are conserved positions defined as identical in over six sequences in at least three species. Where available, 5nt upstream of the 22-bp SL are shown for evaluation of a potential initiator element; sequences consistent with the initiator motif are underlined. Nucleotides existing in some clones but absent in others were italicized. A noncanonical C in the splice-donor site of KbrSL-3 is boxed. The conserved intergenic spacer region between different types of KbrSL are also boxed. Gaps introduced in the sequence alignment are shown as –.
