FIG. 5.—
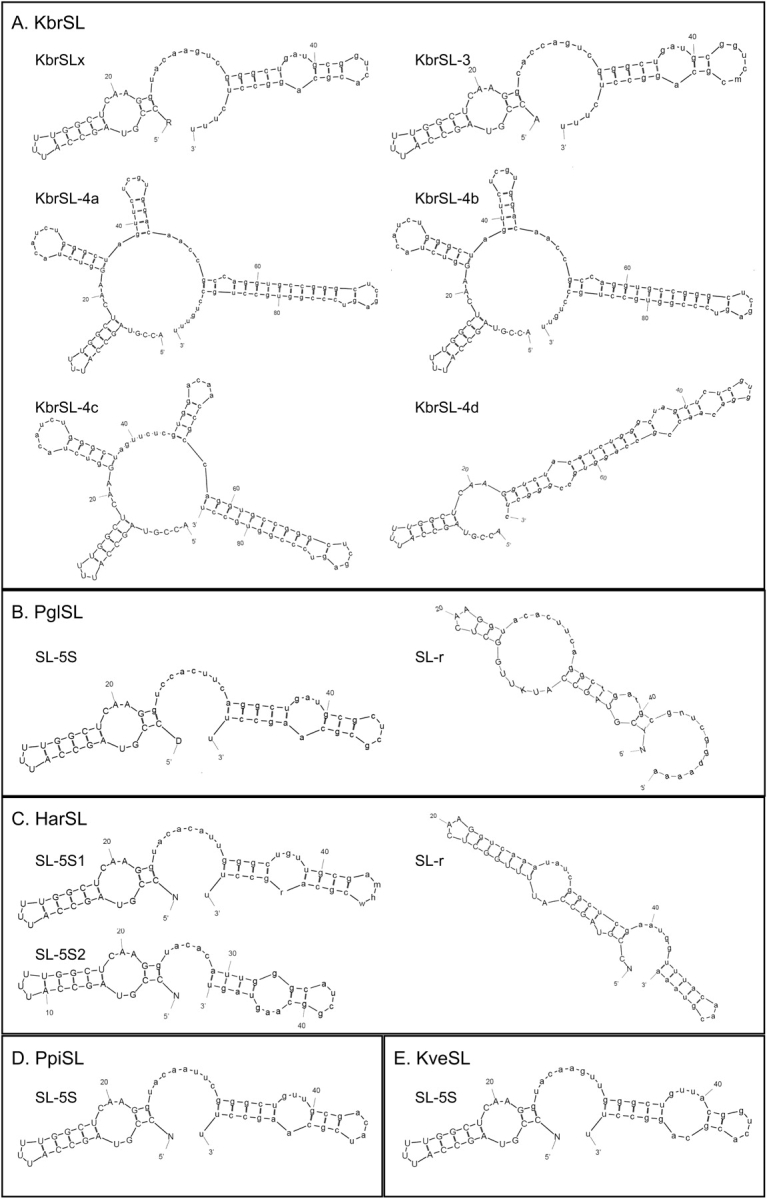
Predicted structures of SL RNA for Karenia brevis (A), Polarella glacialis (B), Heterocapsa arctica (C), and Pfiesteria piscicida (D). Model simulation was run using MFOLD: Prediction of RNA secondary structure modeling program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) under default setting. Simulation for different types of K. brevis SL RNA (KbSL) was based on isolated cDNA and that for P. glacialis (PglSL), H. arctica (HarSL), and the P. piscicida SL–5S form (PpiSL) was based on genomic sequences, by identifying conserved regions in the alignment of SL RNA genes with all the mapped dinoflagellate SL RNA transcripts. SL–5S denotes SL RNA transcribed from the SL–5S genomic cluster; SL-r denotes SL RNA transcribed from genomic tandem repeats of SL RNA gene. For more information, see text.
