Abstract
Purpose
Amplification and deletion of the TOP2A gene have been reported as positive predictive markers of response to anthracycline-based therapy. We determined the status of the HER2 and TOP2A genes in a large cohort of breast cancer patients treated with adjuvant doxorubicin (A) and cyclophosphamide (C).
Patients and Methods
TOP2A/CEP17 and HER2/CEP17 fluorescent in situ hybridization (FISH) were performed on tissue microarrays (TMAs) constructed from 2,123 of the 3,125 women with moderate-risk primary breast cancer who received equivalent doses of either concurrent adjuvant chemotherapy with A plus C (n = 1,592) or sequential A followed by C (n = 1,533).
Results
An abnormal TOP2A genotype was identified for 153 (9.4%) of 1,626 patients (4.0% amplified; 5.4% deleted). An abnormal HER2 genotype was identified for 303 (20.4%) of 1,483 patients (18.8% amplified; 1.6% deleted). No significant differences in either overall survival (OS) or disease-free survival (DFS) were identified for TOP2A. In univariate analysis, OS and DFS rates were strongly and adversely associated only with higher levels of HER2 amplification (ratio ≥ 4.0). Survival was not associated with low-level HER2 amplification (ratio ≥ 2; OS hazard ratio [HR], 1.14; P = .39; DFS HR, 1.07; P = .62), but it was associated for a ratio ≥ 4 (OS HR, 1.45; P = .03; DFS HR, 1.38; P = .033), in which analysis was adjusted for menopausal status, hormone receptor status, treatment, number of positive nodes, and tumor size.
Conclusion
In this population of patients with early-stage breast cancer who were treated with adjuvant AC chemotherapy, TOP2A abnormalities were not associated with outcome. HER2 high-level amplification was a prognostic marker in anthracycline-treated patients.
INTRODUCTION
Results of several clinical trials have documented the improved outcome of patients with breast cancer who were treated in the adjuvant setting with anthracycline-based chemotherapy.1,2 Although anthracycline therapy improves the outcome of treated patients, it is associated with occasional life-threatening toxicities, such as congestive heart failure and acute leukemia, as well as with more common but annoying adverse effects, including nausea and vomiting, mucositis, alopecia, and fatigue. Management of patients in this clinical setting could be enhanced through selective use of these regimens via identification of specific immunophenotypic or molecular markers predictive of response (or absence of response) to the agents employed. Addition of a taxane either concurrently or sequentially to anthracycline-based therapy has been shown to additionally improve patient outcomes,3,4 but this strategy also is encumbered with additional toxicities. Identification of a group of patients who have a low residual risk after treatment with preceding anthracycline-based therapy might spare them the toxicity of requiring subsequent taxane chemotherapy.
Several studies have suggested that amplification and/or overexpression of the ERBB2 (HER2) gene in primary breast cancer tissue may identify a subgroup of patients who are more likely to benefit from anthracyclines than those who have tumors that have normal HER2.5,6 In approximately 35% to 40% of patients with breast carcinoma that demonstrates HER2 gene amplification, topoisomerase II α (TOP2A) is coamplified.7 TOP2A encodes for an enzyme that plays a key role in DNA replication, and it serves as a molecular target for many antineoplastic agents. The gene that encodes TOP2A is located at chromosome 17q 12-q21, in proximity to HER2. Several studies have suggested that, rather than HER2, TOP2A amplification or overexpression is predictive of favorable response to anthracycline-based chemotherapy.8–18 Enigmatically, other reports have demonstrated that both amplification and deletion of TOP2A are related to the sensitivity to anthracycline therapy.19–21 Thus, the simultaneous amplification of HER2 and TOP2A has been proposed as a molecular predictor of response to anthracycline-based regimens.22
In Southwest Oncology Group Protocol S9313 (Intergroup Protocol 0137), patients with either high-risk node-negative or low-risk node-positive breast cancer were randomly assigned to one of two schedules of doxorubicin (A) and cyclophosphamide (C) chemotherapy (combined as AC). Overall results failed to demonstrate any difference in disease-free or overall survival for either of the two tested schedules of AC chemotherapy.23 We hypothesized that patients with TOP2A amplification or deletion would have an outcome superior to patients without such abnormalities when treated with anthracyline-based therapy.
PATIENTS AND METHODS
Patients
Patient selection, assay performance, and data analysis are reported according to the REMARK criteria.24 Tissue microarrays (TMAs) that had been prepared with paraffin blocks collected prospectively from patients who participated in SWOG S9313/Int0137 were used for this study. SWOG S9313 was an adjuvant chemotherapy trial that accrued 3,125 eligible women with early-stage breast cancer from April 1994 through May 1997.23 Participants were required to have one to three nodes involved or to have high-risk node-negative breast cancer, which was defined as primary tumors greater than 2 cm in size or greater than 1 cm for tumors that were both estrogen- and progesterone-receptor negative. Patients were randomly assigned to treatment with one of two alternative dose schedules of AC. As previously reported, there was no difference in disease-free or overall survival for patients treated on the two arms, though the sequential arm (arm 2) produced more myelosuppression and complications related to myelosuppression.23
Construction of TMAs
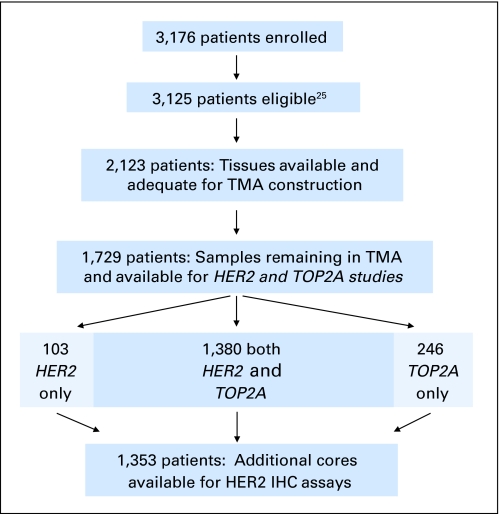
TMAs were constructed from tumor tissue blocks from 2,123 (67%) of the 3,125 patients on S9313.25 Inclusion of tissue for this study is illustrated in Figure 1. All patients provided written informed consent to participate on S9313 as well as to collect blocks for correlative studies.
Fig 1.
REMARK diagram detailing the materials used for this study. Absence of cores, core loss, absence of invasive carcinoma, and insufficient visualization of fluorescent in situ hybridization signals all contributed to absence of scores for cores within the tissue microarrays (TMAs). IHC, immunohistochemistry.
TMA Core Tracking and Fluorescent In Situ Hybridization
Automated TMA core tracking and scoring was performed by using modifications of previously described methods.26 Twenty-eight TMA blocks that contained normal and tumor cores were evaluated by dual-color, direct-label fluorescent in situ hybridization (FISH) by using the TOP2A/CEP17 probe set (Abbott Molecular/Vysis, Des Plaines, IL). Unstained sections from 27 of the TMA blocks were available for evaluation with the HER2/CEP17 probe set by using staining methods previously described.26,27 Absence of cores in TMA blocks, core loss during preparation, absence of invasive carcinoma in cores, and insufficient visualization of signals all contributed to the exclusion of some cores from analysis. The FISH results for available cores that could be scored were averaged, and the mean was used as the FISH score of record.
Automated Scoring of FISH Results
The Metasystems Metafer Metacyte v4.3.1.133 scanning system (Metasystems, Altlussheim, Germany) was used to scan slides and to track TMA cores. The Micro Array Tool (MAT) interface window was used to manually assign each core an identification and to establish coordinate positions for every core, thereby creating a position list for each TMA slide. By using the spot center and relocation functions from the interface window, the core center coordinates were marked and aligned in the overview image and were assigned a core identification from the array map.
Immunohistochemistry
Tissue sections were deparaffinized and rehydrated before they were incubated in 0.01M citrate buffer at pH 6.0 in a steamer for 40 minutes at greater than 95°C. All immunohistochemical procedures were performed on a Dako Autostainer (Dako, Carpinteria, CA). A polyclonal antibody to HER2 (A0485; Dako) was applied at a 1:200 dilution in phosphate-buffered saline (PBS) to sections and was incubated for 40 minutes at room temperature. With intervening wash steps in PBS, slides were incubated for 30 minutes at room temperature in a rabbit-specific, labeled polymer (EnVision+; Dako), which was followed by 10 minutes at 37°C in a solution that contained 3% hydrogen peroxide and 3,3′ -diaminobenzidine. Slides were counterstained with hematoxylin.
Normalized Immunohistochemical Scoring Methodology
Immunostained slides were scored according to a modification of the scoring system approved by the US Food and Drug Administration.28 Only invasive carcinoma was scored among the neoplastic cells. For tumor cells, only membrane staining intensity and pattern were evaluated by using the semiquantitative scale of 0 to 3+. The non-neoplastic epithelium was scored on a scale of 0 to 3+ by using identical criteria. The normalized HER2 score subtracted the score on the benign cells from that on the tumor cells.
Statistical Analysis
Disease-free survival was defined as the time from registration to first recurrence (local, regional, or distant), to new primary cancer in the contralateral breast, or to death as a result of any cause, as per the clinical protocol. Overall survival was defined as time from registration to death as a result of any cause. Patients were censored on the date of last contact if a failure event had not been observed. Unadjusted survival was assessed by the Kaplan-Meier method. Cox regression analysis was used to estimate hazard ratios (HRs) and their 95% CIs; this analysis included possible adjustment for treatment assignment, tumor size (< 2 cm, 2 to 5 cm, or > 5 cm), number of positive lymph nodes (0, 1, 2, or 3), menopausal status, and hormone receptor status (both estrogen- and progesterone-negative status versus either positive by local institutional standards). All reported P values and CIs were from two-sided tests. Statistical testing was done with different cutoffs, as described in the Normalized Immunohistochemical Scoring Methodology section, to determine the sensitivity of the results to the choice of cut point.
To test agreement between immunohistochemical and FISH evaluations of HER2 positivity, we used the κ statistic that corrects for agreement as a result of chance and then determines whether the remaining agreement is significantly greater than chance.29
RESULTS
Patient Demographics
Patients with TMA cores who were included on this study did not differ significantly from those without tissue samples on age, menopausal status, or receptor status. Included patients had slightly larger tumors (P = .049) and slightly more positive nodes (P = .033) than patients without tissue samples. For those with markers, 45% were receptor negative, and 53% had one or more positive nodes. Disease-free survival and overall survival did not differ after adjustment for prognostic factors, so the sample is representative of the entire trial population as described by Linden et al.23
Originally, 2,123 patients (67%) had banked tissue from which adequate TMAs could be constructed.25 After some depletion of the tissue, a total of 11,114 TMA cores from 1,729 patients were sufficiently intact and yielded enumerable FISH signals for scoring (Fig 1). Absence of cores in the TMA blocks, core loss, absence of invasive carcinoma, and insufficient visualization of FISH signals contributed to the exclusion of cores for analysis. HER2 and TOP2A FISH data were available for 1,483 (86%) and 1,628 (94%) women, respectively. The reduced number for HER2 was due to a processing error on one of the 28 TMAs. Both assays could be performed for 1,380 women (80%), whereas the remaining 349 women (20%) had only one assay, not both, completed.
Association of Outcomes With HER2 Abnormalities
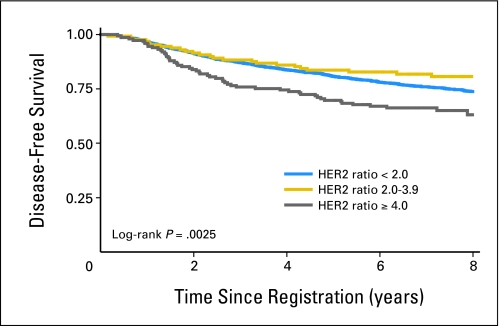
An abnormal HER2 genotype was identified for 303 (20.4%) of 1,483 women (18.8% amplified; ratio ≥ 2.0; 1.6% deleted; ratio ≤ 0.70). Disease-free and overall survival rates were marginally worse with HER2 amplification by classic criteria, but these data were not statistically significant after analysis was adjusted for hormone receptor status, tumor size, number of positive nodes, menopausal status, and randomly assigned treatment (Table 1). Clinical outcomes were not associated with HER2 amplification when considered by consensus agreement for positivity. However, in an exploratory analysis, we observed that both disease-free survival (adjusted HR, 1.38; 95% CI, 1.03 to 1.85) and overall survival (adjusted HR, 1.45; 95% CI, 1.04 to 2.03) were statistically significantly shorter in patients with HER2/CEP17 ratios of greater than 4.0 (Table 1, Fig 2). As the criterion for the HER2 ratio increased, disease-free and overall survival HRs increased, though only those for disease-free survival were statistically significant. No association with survival was detected when patients who had HER2 deletion were compared with those who had normal HER2 or with those who had amplified HER2.
Table 1.
HER2 and TOP2A Gene Amplification Associations
| Genotype | Patient Data |
Disease-Free Survival |
Overall Survival |
|||||
|---|---|---|---|---|---|---|---|---|
| No. | % | HR | 95% CI | P | HR | 95% CI | P | |
| HER2/CEP17 | 1,483 | |||||||
| ≥ 2.0 | 279 | 19 | 1.07 | 0.83 to 1.37 | .62 | 1.14 | 0.85 to 1.52 | .39 |
| ≥ 4.0 | 149 | 10 | 1.38 | 1.03 to 1.85 | .033 | 1.45 | 1.04 to 2.03 | .03 |
| ≥ 6.0 | 51 | 3.4 | 1.76 | 1.14 to 2.73 | .01 | 1.58 | 0.95 to 2.63 | .08 |
| ≥ 8.0 | 16 | 1.1 | 2.48 | 1.27 to 4.82 | .008 | 1.75 | 0.72 to 4.28 | .22 |
| ≤ 0.70 or ≥ 2.0 | 303 | 20 | 1.11 | 0.87 to 1.42 | .39 | 1.21 | 0.91 to 1.60 | .19 |
| TOP2A/CEP17 | ||||||||
| ≥ 2.0 | 65 | 4.0 | 0.98 | 0.60 to 1.59 | .92 | 1.10 | 0.64 to 1.89 | .72 |
| ≥ 4.0 | 13 | 0.8 | 0.52 | 0.13 to 2.08 | .35 | 0.77 | 0.19 to 3.11 | .72 |
| ≥ 6.0 | 4 | 0.3 | NE | NE | NE | NE | NE | NE |
| ≤ 0.70 or ≥ 2.0 | 153 | 9.4 | 1.05 | 0.77 to 1.45 | .75 | 1.24 | 0.87 to 1.78 | .23 |
| TOP2A copy | ||||||||
| ≥ 4.0 | 195 | 12 | 1.07 | 0.80 to 1.44 | .64 | 1.10 | 0.78 to 1.56 | .58 |
| ≥ 6.0 | 48 | 3.0 | 0.70 | 0.38 to 1.32 | .27 | 0.87 | 0.45 to 1.69 | .68 |
| HER2/CEP17 > 4.0 subset | 140 | |||||||
| TOP2A/CEP17 | ||||||||
| ≥ 2.0 | 33 | 24 | 0.79 | 0.40 to 1.59 | .51 | 0.81 | 0.37 to 1.78 | .61 |
| ≥ 4.0 | 7 | 5 | 0.62 | 0.15 to 2.62 | .51 | 0.82 | 0.19 to 3.57 | .79 |
| ≤ 0.70 or ≥ 2.0 | 51 | 36 | 0.78 | 0.43 to 1.41 | .41 | 0.72 | 0.36 to 1.43 | .34 |
NOTE. Analysis was adjusted for hormone receptor status, tumor size, number of positive nodes, menopausal status, and randomized treatment assignment.
Abbreviations: HR, hazard ratio; NE, not estimable.
Fig 2.
The effect of HER2 gene amplification on disease-free survival. Significantly shortened disease-free survival was observed only for high-level amplification (HER2/CEP17 ratio > 4.0).
Association of Outcomes With TOP2A Abnormalities
An abnormal TOP2A genotype was identified in 153 (9.4%) of 1,626 women (4.0% amplified; 5.4% deleted). All but one patient with amplified TOP2A were coamplified for HER2. No significant associations with either disease-free or overall survival were identified for TOP2A amplification and deletion. Both amplification by TOP2A/CEP17 ratio and TOP2A copy number were considered, though neither were associated significantly with survival.
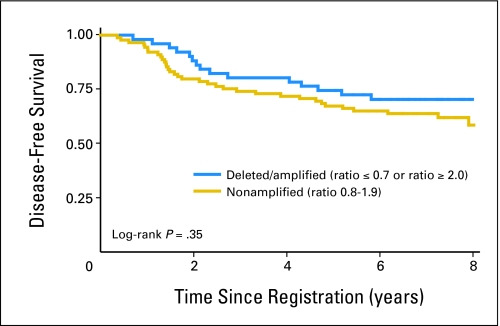
Because TOP2A amplification occurred almost solely in patients with HER2 amplification, we also considered the potential interaction of dual gene amplification—the influence of TOP2A deletion and/or amplification for women with amplified HER2 (ratio ≥ 2.0) and for women who displayed high-level HER2 amplification (ratio ≥ 4.0). As summarized in Table 1 and as illustrated in Figure 3, a statistically significant association was not identified between TOP2A deletion and/or amplification in the context of HER2 amplification, even when patients were segregated by high-level HER2 amplification and by both TOP2A/CEP17 ratio and TOP2A copy number.
Fig 3.
The effect of any TOP2A abnormality (either TOP2A deletion or TOP2A amplification) on disease-free survival for high-level HER2-amplified instances (HER2/CEP17 ratio > 4.0). Similarly, no significant differences in overall survival were observed for TOP2A and HER2 coamplified instances, and neither overall nor disease-free survivals were significantly different when the analysis was segregated for TOP2A deletion or TOP2A amplification within the HER2-amplified group.
Analysis of Outcomes According to Treatment Arm
As noted, there was no difference in disease-free and overall survival in the parent trial in patients randomly assigned to sequential or concurrent AC. In this correlative study, we did not detect any significant differences between these two treatment arms according to HER2 status, even when data were evaluated by using a HER2/CEP17 ratio ≥ 4.0 and TOP2A/CEP17 normal ratio (ie, 0.80 to 1.9; data not shown). Similarly, outcomes in the two arms were similar when analyzed by using HER2/CEP17 ratio ≥ 4.0 and by either TOP2A deletion (ratio ≤ 0.7) or amplification (ratio ≥ 2.0; data not shown).
Correlation of HER2 Amplification by FISH and Immunohistochemical HER2 Status
Tables 2 and 3 summarize the agreement between the HER2/CEP17 ratio and HER2 status by immunohistochemical assay. Among the 845 tumors (62.5%) that stained 0 to 1+ on immunohistochemistry for HER2, 4.1% showed HER2 amplification by FISH. A total of 20.3% of tumors that stained 2+ on immunohistochemistry were found to be HER2 amplified, whereas 80.2% of tumors that stained 3+ on immunohistochemistry showed HER2 amplification by FISH. Statistical comparison of agreement between immunohistochemistry (categorized as 0 to 2+ v 3+) and the HER2/CEP17 ratio (categorized as < 2.0 v ≥ 2.0) showed 89.9% agreement (κ = 0.65; P < .001 for testing against chance agreement).
Table 2.
Correlations of HER2 Determinations by IHC and FISH
| IHC |
HER2/CEP17 Ratio |
|||||
|---|---|---|---|---|---|---|
| < 2.0 |
≥ 2.0 |
Total |
||||
| No. | % | No. | % | No. | % | |
| 0 | 394 | 97.6 | 14 | 3.4 | 408 | 100 |
| 1 | 416 | 95.2 | 21 | 4.8 | 437 | 100 |
| 2 | 236 | 79.7 | 60 | 20.3 | 296 | 100 |
| 3 | 42 | 19.8 | 170 | 80.2 | 212 | 100 |
| Total | 1,088 | 80.4 | 265 | 19.6 | 1,353 | 100 |
Abbreviations: IHC, immunohistochemistry; FISH, fluorescent in situ hybridization.
Table 3.
Correlation of HER2 Determinations by Combined IHC Scores
| IHC |
HER2/CEP17 Ratio |
||
|---|---|---|---|
| < 2.0 | ≥ 2.0 | Total | |
| 0-2 | 1,046 | 95 | 1,141 |
| 3 | 42 | 170 | 212 |
| Total | 1,088 | 265 | 1,353 |
NOTE. IHC was combined into the following categories: 0-2+ and 3+. Statistics were as follows: κ, 0.65; standard error, 0.02; P < .0001.
Abbreviation: IHC, immunohistochemistry.
Association of Outcome With Chromosome 17 Copy Number
Patients who had tumors with aneusomy/polysomy 17 (defined as tumors that had a mean of greater than three CEP17 signals per nucleus) had a significantly better disease-free survival (log-rank P = .016) and overall survival (log-rank P = .028) than did patients with eusomic tumors (defined as having a mean of ≤ three CEP17 signals per nucleus). In an unadjusted Cox model, patients who had tumors with aneusomy/polysomy 17 had significantly better disease-free survival than did patients with eusomy 17 (Cox model HR, 0.84; P = .033 for a one-unit change in CEP17). After analysis was adjusted for receptor status, tumor size, menopausal status, and number of positive nodes, the adjusted HR was 0.86, and P = .075 for a one-unit change in CEP17. For overall survival, an unadjusted Cox model produced an HR of 0.81 and P = .032 for a one-unit change in CEP17. After analysis was adjusted for these same factors, the overall survival HR was 0.83 and P = .071 for a one-unit change in CEP17. When the data were analyzed as described by Reinholz et al,30 in which aneusomy was defined as greater than 30% of nuclei having three or more CEP17 signals, the log-rank P for disease-free survival = .12, though P = .007 if the proportion of aneusomic cells was analyzed as a continuous variable in a Cox regression analysis.
DISCUSSION
In this study, we failed to detect any prognostic effect for classically determined HER2 gene amplification or for TOP2A gene amplification or deletion in tissues collected from a large group of women with modest-risk, stages I and II breast cancer who were all treated with nearly identical adjuvant AC chemotherapy regimens. The correlation between HER2 status as determined by immunohistochemistry and by FISH is similar to that reported by others.29 In an exploratory analysis, we did observe that patients with particularly high levels of HER2 amplification had worse DFS. Reinholz et al30 have previously reported that patients treated with anthracycline- and taxane-based chemotherapy alone (without trastuzumab) whose tumors were HER2 amplified and were polysomic for chromosome 17 had a superior disease-free survival compared with patients with HER2-amplified tumors that were not polysomic for chromosome 17. Too few patients with HER2-nonamplified tumors were present in that group to draw clear conclusions regarding the majority of patients whose tumors are not amplified for HER2. We examined the data in this study in a HER2-unselected population to determine if this finding could be replicated. Although these results are not robust and should not be used to justify the selection of therapy in any patient group, they appear consistent with the report of Reinholz et al.30 Although it is tempting to invoke an explanation for this observation on the basis of topoisomerase II copy number in the tumors with aneusomy/polysomy 17, this explanation is made less tenable by the observations in this study for patients with TOP2A gene amplification, as discussed in the Discussion section; additional studies are needed to additionally explore the relationship between chromosome 17 copy number and outcome and of the biology underlying this apparent association. The analysis was performed by using Cox regression hazard risk adjusted for treatment, menopause, tumor size, the number of positive lymph nodes, and hormone receptor status.
Our results are, at first glance, perplexing. In retrospect, however, we believe they may represent the mixed prognostic and predictive role of HER2 and TOP2A. Several studies have suggested that HER2 amplification is associated with worse prognosis in breast cancer, so one might expect lower disease-free and overall survival in patients with this abnormality.31 Conversely, HER2 and TOP2A appear to be favorable predictive factors for a benefit from anthracycline-containing therapy. Thus, in this population of patients who all received an equivalent regimen of AC, one might expect that patients whose tumors are HER2 and TOP2A positive would have the same outcomes as those whose tumors are negative for these two markers. Their overall prognosis would be worse in the absence of chemotherapy, but they might be expected to achieve more benefit from doxorubicin-based therapy. It has been hypothesized that topoisomerase II abnormalities, which occur almost exclusively in HER2-amplified tumors, are responsible for the previously described association between HER2 amplification and anthracycline sensitivity.8–18 It would be expected that anthracyclines would be of particular benefit in the HER2-amplified tumors that were also amplified or deleted for TOP2A, but not in the HER2-amplified tumors with the normal copy number of TOP2A. The observation that outcome among patients with HER2-amplified tumors did not differ according to TOP2A status suggests that TOP2A amplification or deletion is not the sole explanation for the observation that HER2-amplified tumors benefit from anthracycline-based chemotherapy.
These data do not provide additional insights into how better to treat patients with stages I and II cancer who have modestly high risk for recurrence, if it is assumed that they will receive a doxorubicin-based regimen. Several studies have reported that addition of a taxane to anthracycline-based therapy additionally decreases the risk of recurrence and death,4 although neither increased dose of C or A appears beneficial.4,32,33 A recently reported study from the Cancer and Leukemia Group B (CALGB) has suggested that HER2 amplification and/or overexpression may identify those patients most likely to benefit from addition of pactlitaxel, but there was no detectable interaction between HER2 and D dose greater than 60 mg/m.34 However, in this study, neither HER2 nor TOP2A abnormalities identified a group of patients who might have done so well with AC alone that they would forego paclitaxel. Likewise, with the possible exception of very high HER2 amplification, neither marker distinguished a group of patients whose prognoses appears substantially worse than other groups to the extent that a different treatment strategy would be justified.
HER2 and TOP2A copy number alterations are not associated with outcome in patients who are treated with standard doses of AC given either concurrently or sequentially. Although some studies suggest that these markers might be used to select patients for anthracycline-based therapies,8–17,19–22,35 this finding has not been universal36; because all of the patients on this study were treated with anthracyclines, these data do not bear directly on this question. HER2 is an important predictive marker for selection of anti-HER2–based therapies, such as trastuzumab and lapatinib, and possibly for selection of paclitaxel after adjuvant AC chemotherapy. However, these markers have not been definitively shown to be otherwise useful in selecting additional therapies for patients who receive regimens similar to those received by the patients in this trial.
Appendix
Description of patient materials used in this study.
Tissue microarrays were used for this study that had been prepared using paraffin blocks collected prospectively from patients who participated in SWOG S9313/Int0137. SWOG S9313 was an adjuvant chemotherapy trial that accrued 3,125 eligible women with early-stage breast cancer from April 1994 and May 1997. Participants were required to have one to three nodes involved or to have high-risk node-negative breast cancer, defined as primary tumors less than 2 cm in size, or greater than 1 cm for tumors that were both estrogen- and progesterone-receptor negative. Patients were randomly assigned to treatment with one of two alternative dose-schedules of doxorubicin and cyclophosphamide. Arm 1 consisted of six cycles of treatment with doxorubicin 54 mg/m2 and cyclophosphamide 1.2 g/m2 intravenously every 3 weeks, whereas arm 2 consisted of sequential treatment with four cycles of doxorubicin 40.5 mg/m2 given intravenously on days 1 and 2 of a 3-week cycle, followed by three cycles of treatment with cyclophosphamide 2.4 g/m2 given intravenously every 2 weeks. Radiation therapy was recommended for all women who had breast preservation and for women with mastectomy at the treating physician's discretion. All women with estrogen receptor–positive breast cancer or who were postmenopausal were recommended for adjuvant tamoxifen for 5 years after the conclusion of adjuvant chemotherapy and radiation. As previously reported, there was no difference in disease-free or overall survival for patients treated on the two arms, though the sequential arm (arm 2) produced more myelosuppression and complications related to myelosuppression.
Footnotes
Supported in part by Public Health Service Cooperative Agreement Grants No. CA32102, CA38926, CA49883, CA21155, CA77658, CA25224, CA31946, CA32291, CA37891, CA35431, CA45377, CA58416, CA22433, CA58686, CA46113, CA04919, CA46441, CA58861, CA46282, CA35261, CA27057, CA76132, CA35192, CA76447, CA76462, CA45450, CA76429, CA63845, CA12644, CA20319, CA63844, CA45560, CA58415, CA14028, CA58658, CA42777, CA35119, CA35090, CA35117, CA13612, CA16385, CA67575, CA68183, CA46368, CA04920, CA74647, and CA52654, awarded by the National Cancer Institute, Department of Health and Human Services; by Amgen; and by the Breast Cancer Research Foundation.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Allen Gown, Robert Livingston, Daniel F. Hayes
Administrative support: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Daniel F. Hayes
Provision of study materials or patients: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Peggy Porter, I-Ten Yeh, George Sledge, Charles Shapiro, James Ingle, Charles Haskell, Robert Livingston, Daniel F. Hayes
Collection and assembly of data: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Eric Swain, Allen Gown
Data analysis and interpretation: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Allen Gown, Kathy S. Albain, Robert Livingston, Daniel F. Hayes
Manuscript writing: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Eric Swain, Allen Gown, Daniel F. Hayes
Final approval of manuscript: Raymond Tubbs, William E. Barlow, G. Thomas Budd, Eric Swain, Peggy Porter, Allen Gown, I-Ten Yeh, George Sledge, Charles Shapiro, James Ingle, Charles Haskell, Kathy S. Albain, Robert Livingston, Daniel F. Hayes
REFERENCES
- 1.Levine MN, Pritchard KI, Bramwell VH, et al. Randomized trial comparing cyclophosphamide, epirubicin, and fluorouracil with cyclophosphamide, methotrexate, and fluorouracil in premenopausal women with node-positive breast cancer: Update of National Cancer Institute of Canada Clinical Trials Group Trial MA5. J Clin Oncol. 2005;23:5166–5170. doi: 10.1200/JCO.2005.09.423. [DOI] [PubMed] [Google Scholar]
- 2.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith JA, Ngo H, Martin MC, et al. An evaluation of cytotoxicity of the taxane and platinum agents combination treatment in a panel of human ovarian carcinoma cell lines. Gynecol Oncol. 2005;98:141–145. doi: 10.1016/j.ygyno.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21:976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 5.Pritchard KI, Messersmith H, Elavathil L, et al. HER-2 and topoisomerase II as predictors of response to chemotherapy. J Clin Oncol. 2008;26:736–744. doi: 10.1200/JCO.2007.15.4716. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard KI, Shepherd LE, O'Malley FP, et al. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med. 2006;354:2103–2111. doi: 10.1056/NEJMoa054504. [DOI] [PubMed] [Google Scholar]
- 7.Hicks DG, Yoder BJ, Pettay J, et al. The incidence of topoisomerase II-α genomic alterations in adenocarcinoma of the breast and their relationship to human epidermal growth factor receptor-2 gene amplification: A fluorescence in situ hybridization study. Hum Pathol. 2005;36:348–356. doi: 10.1016/j.humpath.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Di Leo A, Gancberg D, Larsimont D, et al. HER-2 amplification and topoisomerase IIα gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res. 2002;8:1107–1116. [PubMed] [Google Scholar]
- 9.Press MF, Sauter G, Buyse M, et al. Alteration of topoisomerase II-α gene in human breast cancer and its association with responsiveness to anthracycline- based chemotherapy. J Clin Oncol. 2007;25:8s. doi: 10.1200/JCO.2009.27.5644. abstr 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Press MF, Bernstein L, Sauter G. Topoisomerase II-α gene amplification as a predictor of responsiveness to anthracycline-containing chemotherapy in the Breast Cancer International Research Group 006 clinical trial of trastuzumab (Herceptin) in the adjuvant setting. Breast Cancer Res and Treatment. 2005;94:S54. abstr 1045. [Google Scholar]
- 11.Park K, Kim J, Lim S, et al. Topoisomerase II-α (topoII) and HER2 amplification in breast cancers and response to preoperative doxorubicin chemotherapy. Eur J Cancer. 2003;39:631–634. doi: 10.1016/s0959-8049(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 12.Tanner M, Isola J, Wiklund T, et al. Topoisomerase IIα gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24:2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 13.Cardoso F, Durbecq V, Larsimont D, et al. Correlation between complete response to anthracycline-based chemotherapy and topoisomerase II-αgene amplification and protein overexpression in locally advanced/metastatic breast cancer. Int J Oncol. 2004;24:201–209. [PubMed] [Google Scholar]
- 14.Tinari N, Lattanzio R, Natoli C, et al. Changes of topoisomerase IIα expression in breast tumors after neoadjuvant chemotherapy predicts relapse-free survival. Clin Cancer Res. 2006;12:1501–1506. doi: 10.1158/1078-0432.CCR-05-0978. [DOI] [PubMed] [Google Scholar]
- 15.Press MF, Mass RD, Zhou JY, et al. Association of topoisomerase II-α (TOP2A) gene amplification with responsiveness to anthracycline-containing chemotherapy among women with metastatic breast cancer entered in the Herceptin H0648g pivotal clinical trial. J Clin Oncol. 2005;23:847s. abstr 9543. [Google Scholar]
- 16.Di Leo A, Isola J. Topoisomerase IIα as a marker predicting the efficacy of anthracyclines in breast cancer: Are we at the end of the beginning? Clin Breast Cancer. 2003;4:179–186. [PubMed] [Google Scholar]
- 17.Di Leo A, Larsimont D, Gancberg D, et al. HER-2 and topo-isomerase IIα as predictive markers in a population of node-positive breast cancer patients randomly treated with adjuvant CMF or epirubicin plus cyclophosphamide. Ann Oncol. 2001;12:1081–1089. doi: 10.1023/a:1011669223035. [DOI] [PubMed] [Google Scholar]
- 18.Buzdar AU. Topoisomerase IIα gene amplification and response to anthracycline-containing adjuvant chemotherapy in breast cancer. J Clin Oncol. 2006;24:2409–2411. doi: 10.1200/JCO.2006.05.9113. [DOI] [PubMed] [Google Scholar]
- 19.Knoop A, Knudsen E, Balslev E, et al. TOP2A aberrations as predictive and prognostic marker in high-risk breast cancer patients: A randomized DBCG trial (DBCG89D) J Clin Oncol. 2006;24(suppl):11s. abstr 532. [Google Scholar]
- 20.Mano MS, Rosa DD, De Azambuja E, et al. The 17q12-q21 amplicon: Her2 and topoisomerase-IIα and their importance to the biology of solid tumours. Cancer Treat Rev. 2007;33:64–77. doi: 10.1016/j.ctrv.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Knoop AS, Knudsen H, Balslev E, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23:7483–7490. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Järvinen TA, Liu ET. Simultaneous amplification of HER-2 (ERBB2) and topoisomerase IIα (TOP2A) genes–molecular basis for combination chemotherapy in cancer. Curr Cancer Drug Targets. 2006;6:579–602. doi: 10.2174/156800906778742497. [DOI] [PubMed] [Google Scholar]
- 23.Linden HM, Haskell CM, Green SJ, et al. Sequenced compared with simultaneous anthracycline and cyclophosphamide in high-risk stage I and II breast cancer: Final analysis from INT-0137 (S9313) J Clin Oncol. 2007;25:656–661. doi: 10.1200/JCO.2006.07.0847. [DOI] [PubMed] [Google Scholar]
- 24.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 25.Porter PL BW, Yeh IT, Lin MG, et al. P27 (Kip1) and cyclin E expression and breast cancer survival after treatment with adjuvant chemotherapy. J Natl Cancer Inst. 2006;98:1723–1731. doi: 10.1093/jnci/djj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tubbs R. An approach to the validation of novel molecular markers of breas cancer via TMA-based FISH scanning. J Mol Histol. 2007;38:141–150. doi: 10.1007/s10735-006-9076-z. [DOI] [PubMed] [Google Scholar]
- 27.Tubbs RR, Hicks DG, Cook J, et al. Fluorescence in situ hybridization (FISH) as primary methodology for the assessment of HER2 status in adenocarcinoma of the breast: A single institution experience. Diagn Mol Pathol. 2007;16:207–210. doi: 10.1097/PDM.0b013e318064c72a. [DOI] [PubMed] [Google Scholar]
- 28.Gown AM, Goldstein LC, Barry TS, et al. High concordance between immunohistochemistry and fluorescence in situ hybridization testing for HER2 status in breast cancer requires a normalized IHC scoring system. Mod Pathol. 2008;21:1271–1277. doi: 10.1038/modpathol.2008.83. [DOI] [PubMed] [Google Scholar]
- 29.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 30.Reinholz MM, Bruzek AK, Visscher DW, et al. Breast cancer and aneusomy 17: Implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10:267–277. doi: 10.1016/S1470-2045(09)70063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Press MF, Bernstein L, Thomas PA, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: Poor prognosis in node-negative breast carcinomas. J Clin Oncol. 1997;15:2894–2904. doi: 10.1200/JCO.1997.15.8.2894. [DOI] [PubMed] [Google Scholar]
- 32.Fisher B, Anderson S, DeCillis A, et al. Further evaluation of intensified and increased total dose of cyclophosphamide for the treatment of primary breast cancer: Findings from National Surgical Adjuvant Breast and Bowel Project B-25. J Clin Oncol. 1999;17:3374–3388. doi: 10.1200/JCO.1999.17.11.3374. [DOI] [PubMed] [Google Scholar]
- 33.Fisher B, Anderson S, Wickerham DL, et al. Increased intensification and total dose of cyclophosphamide in a doxorubicin-cyclophosphamide regimen for the treatment of primary breast cancer: Findings from National Surgical Adjuvant Breast and Bowel Project B-22. J Clin Oncol. 1997;15:1858–1869. doi: 10.1200/JCO.1997.15.5.1858. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman PA, Broadwater G, Lezon-Geyda K, et al. Correlation of HER2 and chromosome 17 (ch17) copy numbeer with trastuzumab (T) efficacy in CALGB 9840, paclitaxel (P) with or without T in HER2+ and HER2− metastatic breast cancer (MBC) J Clin Oncol. 2007;25:34s. abstr 1009. [Google Scholar]
- 35.Nielsen KV, Ejlertsen B, Moller S, et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta Oncol. 2008;47:725–734. doi: 10.1080/02841860801995396. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett JM, Munro A, Cameron DA, et al. Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 adjuvant breast cancer chemotherapy trial. J Clin Oncol. 2008;26:5027–5035. doi: 10.1200/JCO.2007.14.6597. [DOI] [PubMed] [Google Scholar]