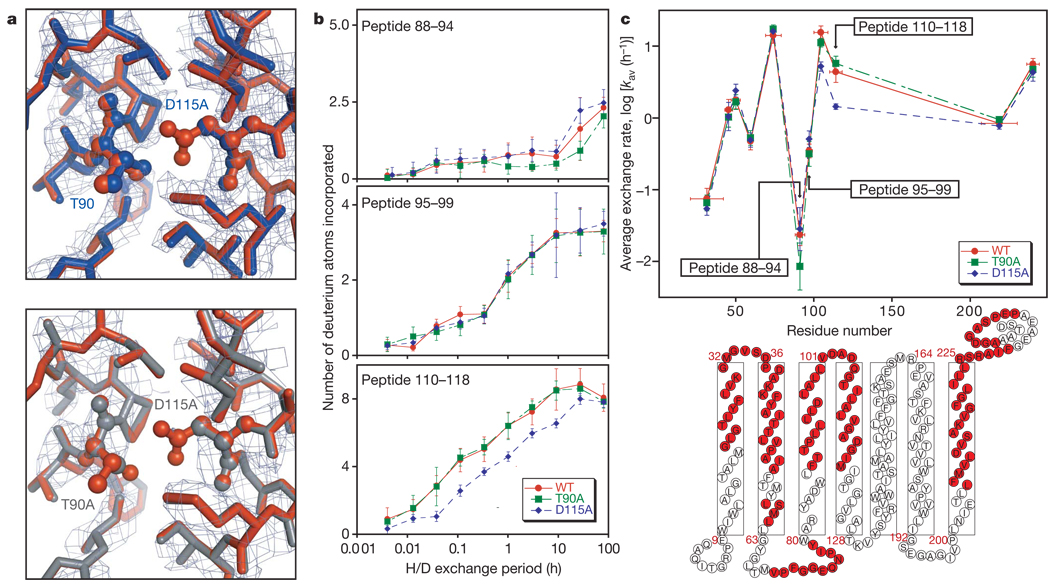
Figure 2. Characterization of the T90A, D115A and T90A/D115Amutants.
a, Omit electron density maps and overlay of refined mutant and wild-type structures for D115A (top) and T90A/D115A (bottom) mutants. The wild-type structure (PDB ID 1PY6) is shown in rust, D115A in blue and T90A/D115A in grey. The mutated side chains are shown in ball-and-stick representation and labelled. The side chains of all residues within 4Å of T90 and D115 of the wild-type (WT) structure were eliminated during refinement for the omit map and are shown here with the exception of W182, which was left out for clarity. The electron density map is contoured at 1.0σ and 1.5σ for D115A and T90A/D115A, respectively. b, Plot of the number of hydrogens exchanged in the denatured state against time for peptides overlapping the T90A mutation (top), a region between T90A and D115A mutation (middle) and the D115A mutation (bottom). In brief, wild-type and mutant proteins were unfolded in SDS and incubated in D2O; the exchange reaction was quenched by rapidly lowering the temperature and pH. The proteins were then digested with pepsin, distinct peptides were separated chromatographically and the change in the mass envelope was measured by electrospray ionization-mass spectroscopy. The maximum scale on the y axis is the maximum number of exchangeable backbone amide hydrogens. Error bars are s.d. estimated with results from triplicate experiments. c, A plot of average exchange rates for peptides throughout the protein (top) and a schematic illustration of the bacteriorhodopsin structure (bottom) showing the sequences covered by the deuterium exchange experiment in light red. Error bars on the x axis reflect the range of the peptic peptides, and those on the y axis are s.d. for ten simulated data sets incorporating the experimental errors observed in the exchange time courses (see Methods).