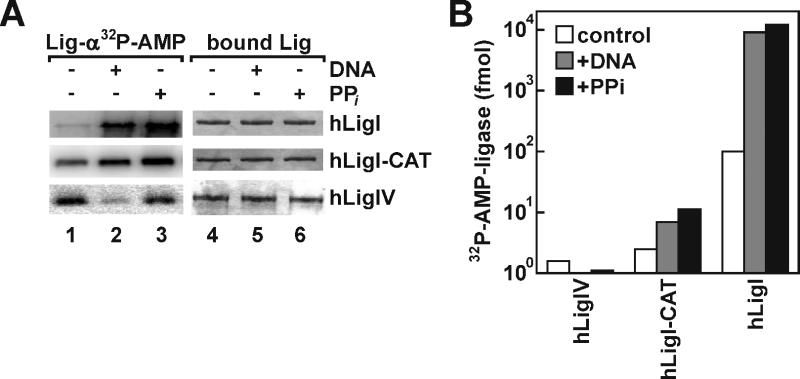
Fig. 5.
Analyses of ligase adenylation. (A) Purified full length hLigI (hLigI), a fragment of hLigI containing the adenylation and OB-fold domains (hLigI-CAT) and hLigIV/XRCC4 (hLigIV) were immobilized on beads as described in Materials and Methods. The immobilized DNA ligases (10 pmol of each) were incubated with: lanes 1 and 4, 10 pmol of [α32P] ATP; lanes 2 and 5, 100 pmol nicked DNA followed by incubation with 10 pmol of [α32P] ATP; lanes 3 and 6, 10 mM pyrophosphate followed by incubation with10 pmol of [α32P] ATP as described in Materials and Methods. After elution from beads, proteins were separated by SDS-PAGE. Total proteins were detected by Coomassie blue staining (right panel) and labeled, adenylated proteins were detected and quantitated by PhosphorImager analysis (left panel). (B) Graphical representation of the ligase-adenylation assay. The horizontal axis numbers correspond to the lane designations in (A). The amount of labeled adenylated ligase was determined by liquid scintillation counting of the radioactive band.