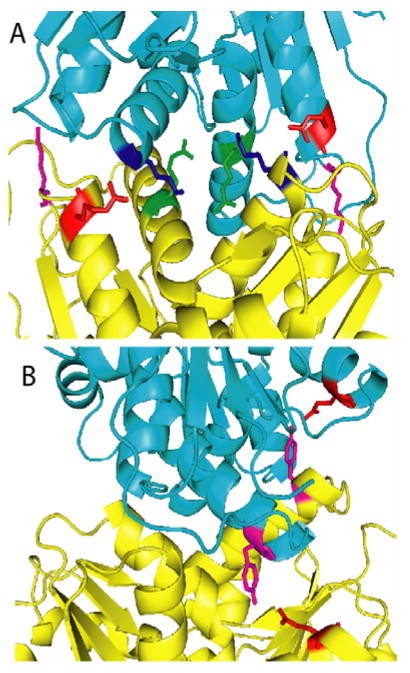
Figure 6.
(A) Crystal structure of RacE2. The two monomers are shown in different colors (yellow and cyan). The residues at the dimer interface that were mutated to Ala are indicated. R25 (red) interacts with the backbone of K106 (blue). K29 (magenta) is located on a large loop on either side of the dimer interface and interacts with the side chain of Asn 136 and the backbones of Lys132 and Ser133 (not shown). R214 (green) is located in the center of the dimer interface and interacts with the side chains of T103, R215 and the backbone of P99 (not shown). (B) Q86 (red) and Y221 (magenta) are both located on one face of the dimer interface with Q86 interacting with the side chain of Y221 from the other monomer. This figure was generated by PyMOL.