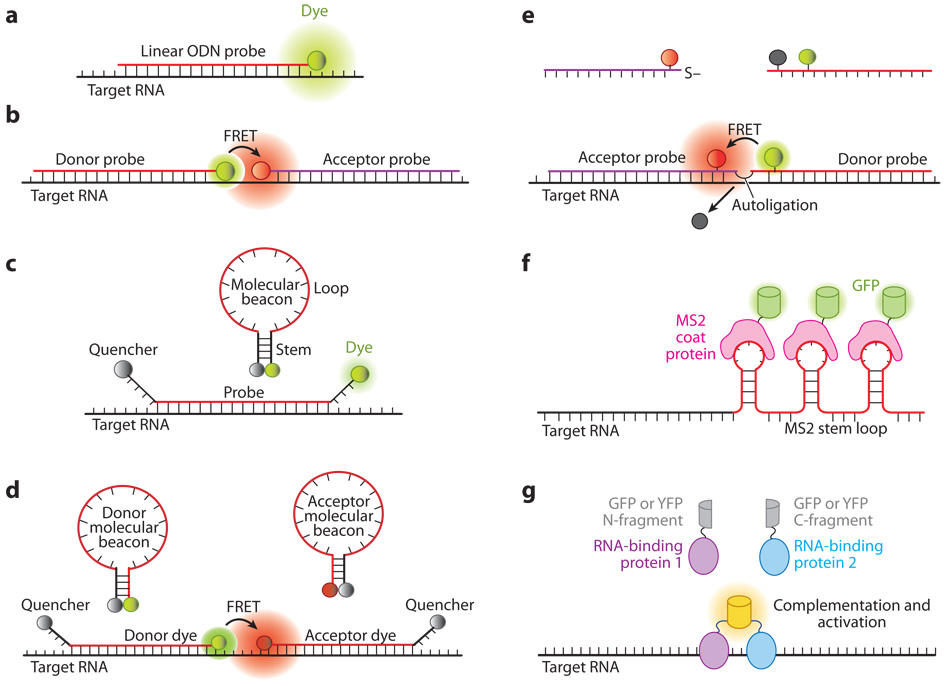
Figure 1.
Illustrations of fluorescent probes for live-cell RNA detection. (a) Tagged linear oligonucleotide (ODN) probes. (b) Linear fluorescence resonance energy transfer (FRET) probes in which two linear ODN probes have, respectively, a donor and an acceptor fluorophore that form a FRET pair. (c) Molecular beacons are dual-labeled stem-loop oligonucleotide hairpin probes with a reporter fluorophore at one end and a quencher molecule at the other end. (d) Dual FRET donor and acceptor molecular beacons hybridize to adjacent regions on the same mRNA target, resulting in FRET signal. (e) Autoligation FRET probes. The fluorescence of the donor is initially quenched. Upon binding of the two probes to adjacent sites on the same RNA, the quencher is displaced and the ligation brings the donor and acceptor fluophores together, resulting in FRET signal. ( f ) Probes using the coat protein of the bacterial phage MS2 fused with green fluorescent protein (GFP) (MS2-GFP). The MS2-GFP complexes bind to multiple hairpin sequences in the 3′ untranslated region (UTR) of an mRNA, giving rise to a high signal compared with the background. (g) Probes based on fragment complementation of fluorescent protein. When two RNA-binding proteins, each carrying a fluorescent protein fragment, bind to adjacent sites on the same RNA, a fluorescence signal is generated owing to fragment complementation of the fluorescent protein.