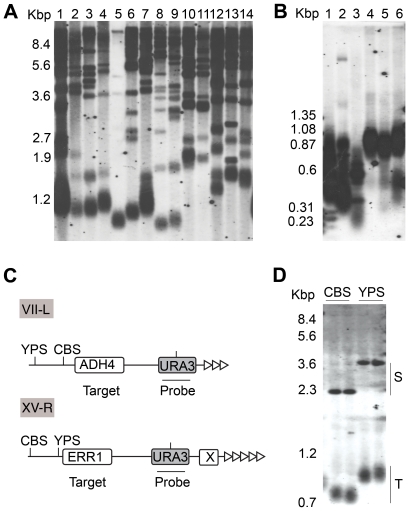
Figure 1. Telomere length screening in Saccharomyces strains.
(A) XhoI digestion of genomic DNA of several S. cerevisiae and S. paradoxus strains probed with telomeric TG1–3 repeats. S. cerevisiae: 1) S288c, 2) DBVPG6763, 3) Y55, 4) SK1, 5) DBVPG6693, 6) YPS128, 7) DBVPG1108; S. paradoxus: European, 8) CBS432, 9) N-17; Far Eastern, 10) NBRC1804, 11) N-44; North American, 12), YPS125, 13) YPS138, 14) DBVPG6304. (B) Genomic digestion using enzymes that do not digest within telomeric repeats. S. cerevisiae: 1) S288c, 2) Y55; S. paradoxus: 3) CBS432, 4) N-44, 5) N-45, 6) N-46. (C) Single telomere tag of chromosomes VII-L and XV-R. Insertion point of the URA3 is indicated (target). Probes (horizontal line) and ScaI restriction sites (vertical line) are indicated. (D) Southern blot of genomic DNA digested with ScaI and probed with URA3. Two independent transformants of CBS and YPS tagged at the VII-L telomere and propagated for 250 generations are shown. S indicates the internal fragment and T the fuzzy telomere end. Subtelomeric polymorphic restriction sites generate different restriction fragments in the YPS and CBS strains.