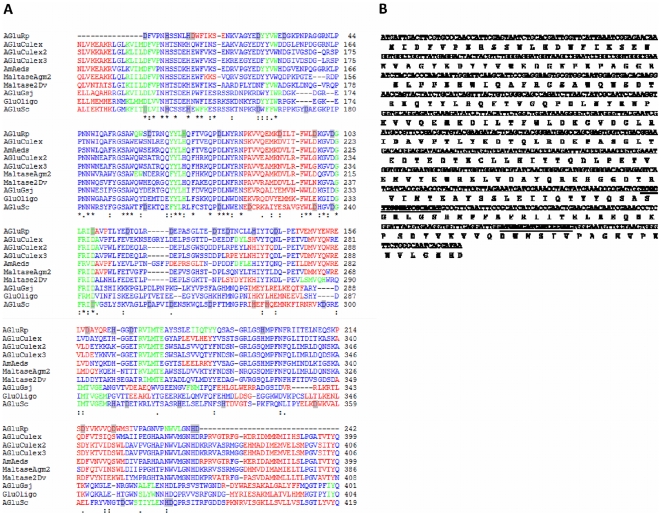
Figure 5. Analysis of the α-glucosidase sequence.
A. Alignment of amino acid sequences of α-glucosidase from R. prolixus (AgluRp), Culex quinquefasciatus (AGluCulex, AGluCulex2, AGluCulex3), α-amylase from Aedes aegypti (AmAeds), maltase-like Agm2 from Anopheles gambiae (MaltaseAgm2), maltase 2 from Drosophila virilis (Maltase2Dv), α-Glucosidase from Gsj (AgluGsj), Oligo-1,6-Glucosidase from Bacillus cereus (GluOligo) and α-glucosidase from Saccharomyces cerevisiae (AgluSc). Identical residues are indicated by “*”; conserved and semiconserved residues are indicated by “:” and “.”, respectively. Residues of aspartic acid and histidine present in AGluRp and also present in AGluSc are marked in gray. The secondary structure prediction using the JPred server is represented in red (α-helices), green (β-sheets) and blue (loops). B. Partial nucleotide sequence of the R. prolixus α-glucosidase cDNA and its deduced amino acid sequence. The amino acid sequences used for the design of specific Real Time-PCR primers are underlined.