Fig. 3.
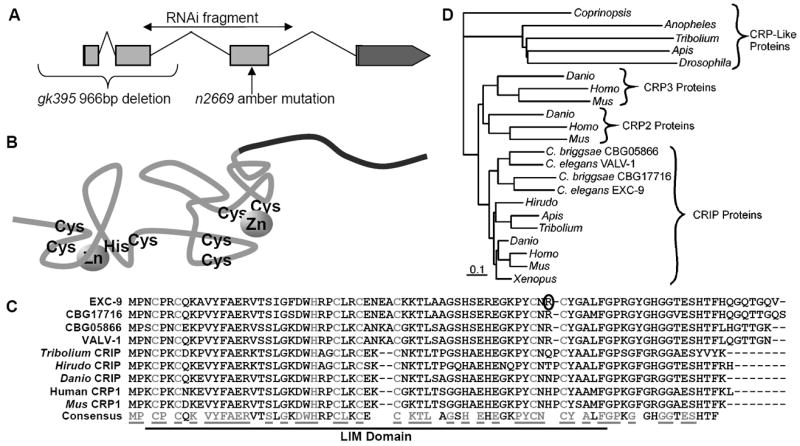
EXC-9 is a conserved small LIM-domain protein, CRIP. (A) Structure of the exc-9 gene, indicating exons, introns, and UTRs. The position of the n2669 and gk395 mutations are indicated, as is the amplified region used for PCR in order to perform RNAi. (B) exc-9 encodes a small protein containing a single LIM domain with 2 Zinc-finger domains, plus a short variable-length and -shape tail (in black) at the C-terminus. Structure (adapted from (Perez-Alvarado et al., 1996)) shows approximate positions of conserved cysteine and histidine residues used to co-ordinate zinc binding. (C) Alignment of EXC-9 and other CRIP/CRP1 proteins shows well-conserved homology. The Zn-binding cysteine and histidine residues of the LIM domain are shown in grey, as are all residues in the consensus sequence that are 100% conserved between these proteins. The arginine residue of EXC-9 that is mutated to a stop codon in the n2669 allele is circled. (D) Phylogenetic tree of EXC-9 and CRIP homologues suggest that EXC-9 and VALV-1 result from a duplication found only in nematodes. EXC-9 and VALV-1 show closest homology to the CRIP/CRP1 family of vertebrate proteins.
