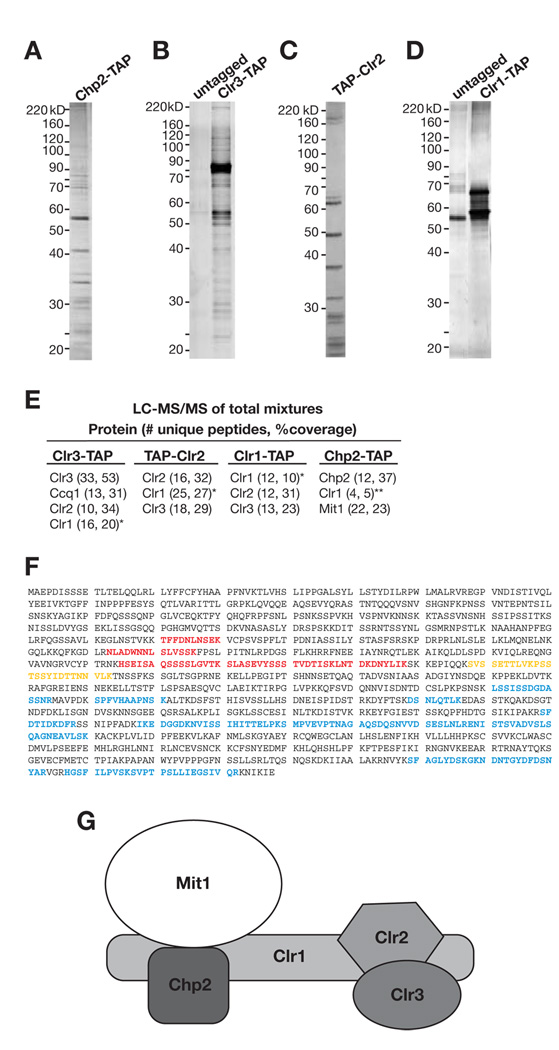
Figure 1. Purification of Chp2 and identification of the SHREC2 complex.
Representative silver-stained gels depicting the tandem affinity purifications of (A) Chp2-TAP, (B) Clr3-TAP, (C) TAP-Clr2, and (D) Clr1-TAP proteins. (E) Proteins identified by tandem mass spectrometry sequencing of mixture of proteins (LC-MS/MS) for the indicated purification. The numbers in parentheses correspond to the number of unique peptides and protein coverage based on total number of amino acids. (F) Clr1 peptide spectra in SHREC2 purifications. Red, residues represent peptides only identified in Chp2-TAP purification. Blue residues represent peptides identified in Clr3-TAP, TAP-Clr2 or Clr1-TAP purifications. Yellow residues show peptides common to both Chp2-TAP and TAP-Clr2 purifications. (G) SHREC2 complex is composed of Chp2 and Mit1, which interact with the N-terminal region of Clr1, and Clr2 and Clr3, which interact with the C-terminal region of Clr1.