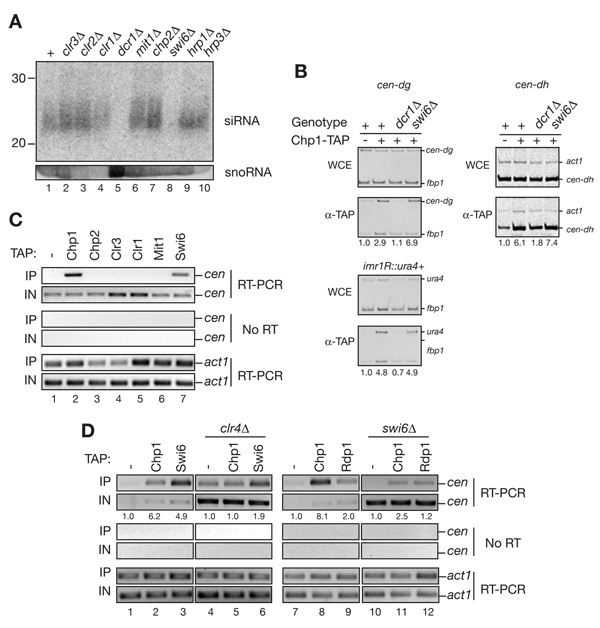
Figure 6. Swi6, but not SHREC2, associates with noncoding cenRNAs and is required for their processing into siRNAs.
(A) Northern blot showing that Swi6 is required for accumulation of centromeric siRNAs. Size fractionated total small RNAs purified from the indicated cells were probed with end-labeled DNA oligos complementary to centromeric siRNAs. The membrane was then re-probed with snoR69, serving as a loading control. Unlike swi6Δ, SHREC2 mutant cells exhibited no defect in siRNA production. (B) ChIP experiments showing that RITS recruitment to centromeric dg, dh, and imr1::ura4+ is Swi6 independent. Immunoprecipitations were performed using wild-type, dcr1Δ, and swi6Δ cells carrying a chp1-TAP gene. Untagged cells were used as a control. Values below each lane represent enrichment compared to the untagged control fbp1+ and act1+ were used as internal controls. (C) RNA immunoprecipitation (RNA-IP) experiments showing that centromeric transcripts (cen) associate specifically with Swi6, but not with SHREC2 components Chp2, Clr3, Mit1 or Clr1. Chp1 is used as a positive control. Actin mRNA (act1) was used as a control to show that the interaction is specific to heterochromatic transcripts. (D) RNA-IP experiments showing that Chp1 and Swi6 associate with centromeric transcripts in a Clr4-dependent manner (lanes 1–6); efficient Chp1 and Rdp1 interaction with cen transcripts requires Swi6 (lanes 7–12). Actin mRNA (act1) and the untagged sample were used as specificity controls. The number shown below each lane is the average enrichment calculated by comparing the enrichment values to the untagged control in two independent experiments.