SUMMARY
The history of life offers plentiful examples of convergent evolution, the independent derivation of similar phenotypes in distinct lineages [1]. Convergent phenotypes among closely related lineages (frequently termed “parallel” evolution) are often assumed to result from changes in similar genes or developmental pathways [2], but the genetic origins of convergence remains poorly understood. Ninespine (Pungitius pungitius) and threespine (Gasterosteus aculeatus) stickleback fish provide many examples of convergent evolution of adaptive phenotypes, both within and between genera. The genetic architecture of several important traits is now known for threespine sticklebacks [3–10]; thus, ninespine sticklebacks thus provide a unique opportunity to critically test whether similar or different chromosome regions control similar phenotypes in these lineages. We have generated the first genome-wide linkage map for the ninespine stickleback and used quantitative trait locus (QTL) mapping to identify chromosome regions controlling several skeletal traits and sex determination. In ninespine sticklebacks, these traits mapped to chromosome regions not previously known to control the corresponding traits in threespine sticklebacks. Therefore, convergent morphological evolution in these related, but independent, vertebrate lineages may have different genetic origins. Comparative genetics in sticklebacks provides an exciting opportunity to study the mechanisms controlling similar phenotypic changes in different groups of animals.
RESULTS AND DISCUSSION
Genome-wide linkage map
The last several years have witnessed substantial progress in characterizing the genetic basis of adaptive diversity in natural populations and species. In threespine sticklebacks, development of new genetic and molecular tools has made it possible to identify major loci controlling repeated evolution of armor plate, pelvic, opercular, and skin color changes in populations that colonized new lakes and streams generated by widespread deglaciation beginning about 20,000 years ago [3–6, 9, 10]. An emerging theme of genetics studies in threespine sticklebacks is that the same genes or chromosome regions underlie similar phenotypes in multiple natural populations, including major effects of Pitx1 [4, 6, 11, 12] and Ectodysplasin (Eda) [5, 6, 8] in the evolution of derived pelvic and armor phenotypes, respectively, throughout the range of this species. Development of comparable genetic resources for ninespine sticklebacks makes it possible for us to critically compare the genetic basis of convergent evolution in a fish group that has also evolved a number of similar interesting morphological and physiological changes (Fig. 1), but last shared a common ancestor with threespine sticklebacks well over 13 million years ago [13].
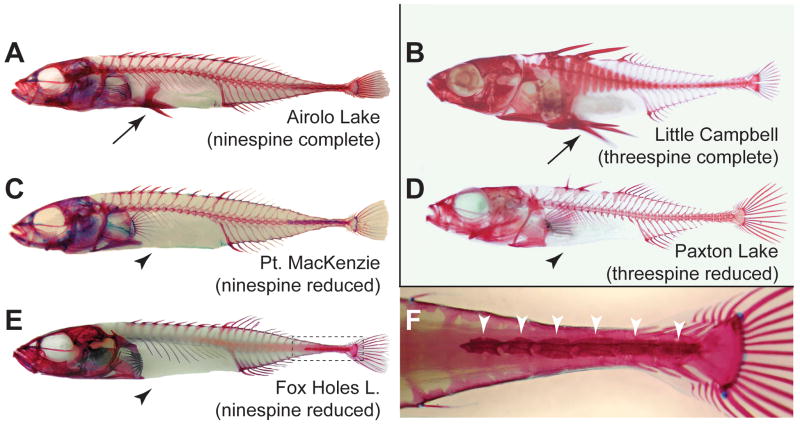
Figure 1. Convergent skeletal evolution in ninespine and threespine sticklebacks.
Reduction and loss of the pelvic (hind) fin has evolved in multiple populations of both ninespine and threespine sticklebacks. (A,B) Ninespine (A) and threespine (B) sticklebacks with complete pelvic skeletal structures (arrow) from Airolo Lake, Alaska; and Little Campbell River, British Columbia. (C,E) Ninespine sticklebacks missing all pelvic structures (arrowhead) from Point MacKenzie, Alaska; and Fox Holes Lakes, Northwest Territories. These two populations were used in the mapping cross. (D) A similar pelvisless phenotype occurs in the benthic threespine sticklebacks of Paxton Lake, British Columbia. (F) Enlargement of boxed area in E showing detail of caudal portion of bony armor (arrowheads), which varies in numbers of plates among fish from different populations and in our laboratory cross. All specimens were cleared by digestion in trypsin and stained in alizarin red S to visualize ossified skeletal structures. Photographs are not to scale.
To generate a genome-wide linkage map for quantitative trait locus (QTL) studies, we produced a Pungitius pungitius genomic library, screened it with a probe for microsatellite repeats, sequenced individual clones, and designed PCR primers that could amplify individual microsatellite repeat regions from ninespine stickleback genomic DNA samples. We typed 212 microsatellite markers (169 derived from ninespine sticklebacks and 43 from threespine sticklebacks) on 120 F1 progeny from a cross between Canadian and Alaskan ninespine sticklebacks, both lacking pelvic structures (Fig. 1C, E). The female parent came from Fox Holes Lakes (Northwest Territories, Canada), which is monomorphic for total absence of the pelvis [14]. The male parent came from an unnamed creek on Pt. MacKenzie (Matanuska-Susitna Borough, south-central Alaska), where ninespine sticklebacks are polymorphic for pelvic phenotypes (average pelvic score for the Pt. MacKenzie population is 1.96 on a scale [15] that ranges from 0 (bilateral absence of pelvic structures) to 8 (all 4 pelvic elements present on both sides)). The combined ninespine and threespine markers defined 30 genetic linkage groups (LGs), comprising 190 markers (151 from ninespine sticklebacks, 39 from threespine sticklebacks) and spanning a total genetic distance of 957.8 cM (Supplemental Figure 1). Since cytological studies show that Pungitius has 21 chromosomes [16–18], we expect some current linkage groups to coalesce with others as additional markers are added to the map.
To compare LGs in ninepsine and threespine sticklebacks, we examined map locations of 39 markers that could be amplified from genomic DNA in both species. We also used BLAST searches to compare the unique sequences from the 151 newly isolated and mapped ninespine stickleback markers with an initial genome assembly for the threespine stickleback (http://www.ensembl.org/Gasterosteus_aculeatus/index.html). In all, 88.7% (134/151) of ninespine stickleback marker sequences could be mapped to unique threespine stickleback chromosome scaffolds, 2.0% (3/151) mapped to unassembled scaffolds, and the remaining 9.3% (14/151) of sequences either produced no significant BLAST results or mapped to multiple genomic scaffolds (Supplemental Table 1). At least 50% of markers in each ninespine linkage group were associated with a single threespine chromosome (87.9% of markers overall, mean of 85.2% of markers per linkage group). These results suggest that synteny has been well conserved between the two genera, both of which have 21 cytologically visible chromosomes [16]. For ease of comparison of results between species, we numbered linkage groups in the ninespine genetic map to match the syntenic linkage group in the threespine map.
Comparative mapping of pelvic reduction
A dramatic example of convergent evolution between populations and genera of sticklebacks is the reduction or loss of the pelvic (hind fin) skeleton (Fig. 1). The pelvis is present in all marine and most freshwater populations of threespine and ninespine sticklebacks, but it has been lost repeatedly in several freshwater populations, likely as an adaptation to local predators and water chemistry [4, 14, 15, 19–24]. Previous studies of threespine sticklebacks have identified one QTL of major effect on LG7 that controls more than 50% of the variation in pelvic size in crosses from diverse geographic locations, including British Columbia, Alaska, Iceland, and Scotland [4, 6, 12]. Mapping, sequencing, and expression studies suggest that this major QTL corresponds to the Pitx1 locus [4, 12, 25], a homeodomain transcription factor that is expressed in developing hindlimbs but not forelimbs of vertebrates [26–28]. Previous complementation and in situ studies show that Fox Holes Lakes ninespine sticklebacks have recessive genetic changes that also reduce Pitx1 expression in the pelvis [29].
Presence or absence of a pelvic skeleton segregates in a 1:1 Mendelian ratio in our ninespine stickleback cross (Fig. 2). Of the 120 progeny analyzed, 59 had complete pelvic skeletons (bilateral presence of an anterior process, posterior process, ascending branch, and spine [30]), 8 had partial skeletons (6 of these had fewer than half of the normal structures), and 53 lacked all pelvic structures. The binary, qualitative trait of presence versus absence of the pelvic complex (partial phenotypes excluded) mapped to LG4 with a peak LOD score of 82.16 (Fig. 2D, Supplemental Tables 2 and 4). Detailed analysis of marker genotypes shows that the striking dimorphism in this cross originates from the Alaskan male parent – inheritance of one Alaskan parental LG4 haplotype is usually associated with a complete pelvis in the F1 progeny, while inheritance of the other Alaskan haplotype is usually associated with the absence of pelvic structures (Supplemental Table 4). Thus, the Alaskan male parent of the cross, which comes from a population that is polymorphic for pelvic phenotypes, was heterozygous for a dominant allele for pelvic reduction. The phenotypic effect of the LG4 region in the current cross is as large as that reported previously for the Pitx1 (LG7) region in threespine sticklebacks [4, 6, 12], but maps to a completely different linkage group. This QTL on LG4 is unlinked to a marker in an intron of the ninespine stickleback Pitx1 gene (Pun319), and to two markers on threespine stickleback BAC clones containing the Pitx1 gene (Stn430, Stn431; Fig. 2E and Supplemental Fig. 1). The region of the threespine stickleback genome that corresponds to the pelvic reduction region in the ninespine contains several genes with known roles in limb and fin development, including members of the Fgf, Msx, and Wnt families. We are currently investigating the potential roles of these candidate genes in pelvic reduction in the Point MacKenzie population.
Figure 2. Pelvic reduction maps to LG4, not to Pitx1, in a ninespine stickleback cross.
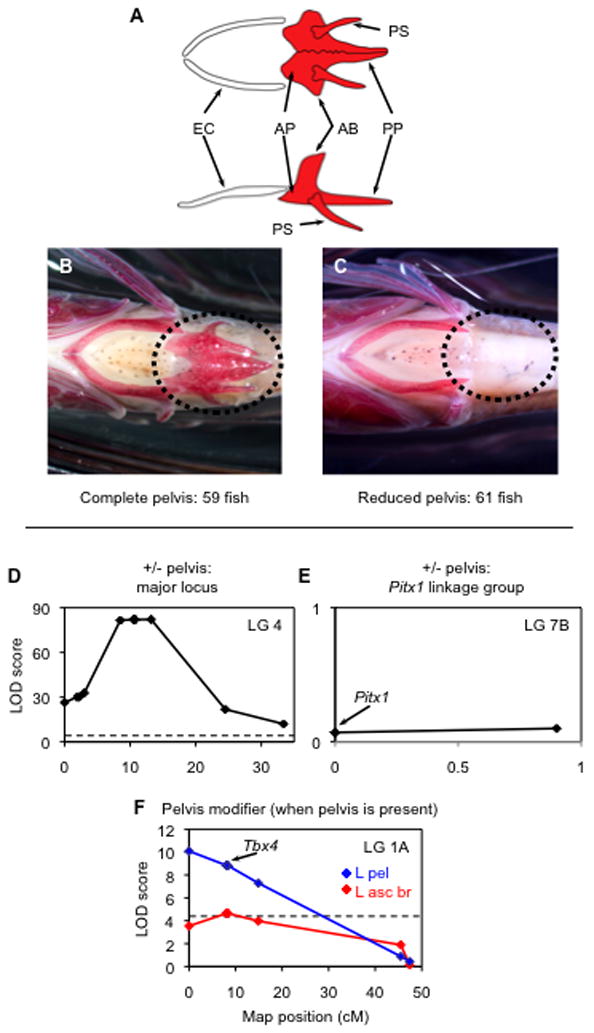
(A) Morphology of the ninespine stickleback pelvis and ectocoracoid in ventral (top) and lateral (bottom) views. A complete pelvis shows bilateral presence of the anterior process (AP), posterior process (PP), ascending branch (AB), and pelvic spine (PS). Pelvic girdle length was measured from the anterior tip of AP to the posterior tip of PP. Anterior to the pelvis is the ectocoracoid bone (EC) of the pectoral girdle. Diagrams modified after [30]. (B,C) The 120 progeny showed a 1:1 ratio of (B) complete to (C) reduced pelvic phenotypes. Anterior is to the left in both images. (D) A QTL on LG4 controlled presence versus absence of the pelvis. Only informative markers (polymorphic in the Alaskan male parent) are shown. The plateau of the LOD peak is due to low recombination between LG4 haplotypes in the Alaskan parent of the cross. (E) The linkage group containing Pitx1 did not have a significant effect on pelvic phenotype. (F) Restricted MQM analysis detected an additional QTL interval influencing left ascending process height (L asc pr; red) and pelvic girdle length (L pel; blue), and this interval includes the Tbx4 gene, a transcription factor involved in hindlimb development [27]. Dashed lines: LOD significance threshold (95% genome-wide level of ≥ 4.5 in D [58] and ≥4.3 in F; not shown in E to limit LOD scale preserve visibility of plot).
Ninespine sticklebacks from the Pt. MacKenzie, Alaska, population show a key morphological difference compared to most other pelvic reduced populations. Most extant and fossil threespine stickleback populations [31], mice with knockouts in the Pitx1 gene [32], and Florida manatees with vestigial pelvic structures [29] show greater pelvic reduction on the right than left side. In contrast, the Pt. MacKenzie ninespine sticklebacks tend to show greater pelvic reduction on the left than right side. Our linkage studies provide the first genetic evidence that populations with different types of directional asymmetry have changes in different major genes controlling pelvic reduction. Approximately 10% of threespine stickleback populations with extensive pelvic reduction show greater reduction on the left than right side [31]. It will be interesting to see if pelvic reduction in these populations maps to the same region detected in Pt. MacKenzie ninespine sticklebacks.
Although the major QTL for pelvic reduction in our ninespine cross is clearly distinct from the Pitx1 locus, the position of the QTL on LG4 is in a region similar to a pelvic modifier QTL that controls less than 6% of the variation in pelvic spine length and pelvic girdle length in a cross between marine (complete pelvis) and pelvic-reduced threespine sticklebacks [4] (Table 1). It is possible that similar genes contribute to pelvic reduction in both threespine and ninespine sticklebacks, but the magnitude of their phenotypic effects differs dramatically between genera. The large impact of the LG4 region in the ninespine fish likely depends in part on a sensitized genetic background in the Pt. MacKenzie by Fox Holes Lakes cross, where all F1 progeny also inherited pelvic reduction alleles from the Fox Holes Lakes parent [29]. The LG4 region in the current cross has a larger phenotypic effect than either Ectodysplasin (Eda,) Pitx1, or Kit ligand (Kitlg) genes in threespine sticklebacks, each of which has been successfully isolated by mapping or positional cloning studies [4, 8, 10]. Ninespine sticklebacks should thus provide a very useful system for identifying additional loci controlling major evolutionary phenotypes in natural populations.
Table 1.
Comparison of QTL for skeletal traits and sex determination in ninespine and threespine sticklebacks (similar mapping results highlighted in bold italics).
| Trait | Ninespine LG | Threespine LG | References |
|---|---|---|---|
| Pelvis (complete vs. reduced) | 4 | 7 | [4, 6, 12] |
| Ascending branch height | 1, 4 | 7, 10 | [4] |
| Pelvic girdle length | 1, 4 | 1, 2, 4, 7 | [4] |
| Pelvic spine length | 4 | 2, 4, 7, 8 | [3, 4] |
| Lateral plate number | 12 | 4, 7, 10, 26* | [5, 6, 8] |
| Sex determination | 12 | 19 | [7] |
Chromosome 21 in threespine stickleback genome assembly.
A single region on ninespine LG4 largely controls the presence-versus-absence pelvic phenotype, yet other chromosome regions control quantitative variation in pelvic size in those progeny that do have a pelvis. For example, we identified a region on LG1A that controls up to 33.2% of the variation in left and right pelvic structures, with a more pronounced effect on the left than the right side (Fig. 2, Supplemental Fig. 1, Supplementary Tables 2 and 4). The LG1A QTL in the ninespine cross overlaps the broad location of a QTL interval that controls approximately 6% of the variance in pelvic girdle length in a threespine stickleback cross [4].
Notably, variation at the major and modifier pelvic loci reveals cryptic genetic variation (CGV) in the wild Pt. MacKenzie population. CGV is thought to be an important and pervasive, yet underappreciated, factor in the response of organisms to mutation, selection, and disease [33]. Both parents of our cross had similar pelvisless phenotypes, yet half of their progeny developed complete pelvises on the hybrid genetic background. Most of the fish in the wild Pt. MacKenzie population also exhibit extreme pelvic reduction, so much of the variation in the major and modifier pelvic loci may remain hidden except under extreme environmental conditions, or in cases of genetic perturbations such as hybridization with other genetic backgrounds, as is the case on our cross (reviewed in [33]). This study provides a dramatic example of the phenotypic diversity that can result when admixture occurs between different outbred genetic backgrounds.
A novel chromosome region controls lateral armor in ninespine sticklebacks
Other skeletal traits mapped to different regions of the genome in ninespine sticklebacks relative to threespine sticklebacks (Table 1, Supplemental Fig. 1, Supplemental Tables 2–5). In threespine sticklebacks, lateral plate variation maps to the Eda locus on LG4 [5, 6, 8]. We mapped two markers in and around the Eda locus in our ninespine cross, including Stn364 (located in an intron of the Eda gene itself), and the closely linked Stn361 marker (located 16 kb away, just 5′ of the Eda locus). Both markers mapped to LG4 in the ninespine stickleback cross, in a chromosome region that did not have significant effects on plate phenotypes. Instead, lateral plate number variation in the ninespine stickleback cross (Fig. 1F) mapped to LG12, the same chromosome region that determines sex (see below). This linkage group accounted for nearly one-third (30.1% left-side and 28.4% right side) of the variance in plate number. Notably, unlike the major pelvic locus on LG4, segregation of different alleles on LG12 from both the Alaskan and Northwest Territories parents had significant effects on plate phenotypes (Supplemental Tables 3 and 5). The armor QTL on LG12 is also distinct from all known chromosome regions that have smaller quantitative effects on armor phenotypes in threespine sticklebacks with reduced numbers of plates (Table 1).
The sex determination locus differs between stickleback genera
Several different mechanisms underlie sex determination among teleost fishes, and under normal conditions sex can be determined by genetic and/or environmental cues [34, 35]. Sex determination in threespine sticklebacks behaves as a simple Mendelian trait that maps to LG19 [7]. Sex determination in ninespine sticklebacks also behaves as a Mendelian trait, but it maps to LG12, in a completely different region of the genome relative to markers closely linked to the sex-determining region in threespine sticklebacks (Stn186, Stn194) [7] (Table 1, Supplemental Fig. 1, Supplemental Tables 3 and 5).
The sex determination region of LG19 in threespine sticklebacks shows striking differences in recombination rates in male versus female meiosis [7]. Similarly, LG12 in the ninespine stickleback cross covers approximately 13 cM, due largely to a lack of recombination in male meioses (Supplemental Figs. 1 and 2). In contrast, when female meioses were analyzed independently of male meioses, the genetic distances between markers are greater and LG12 covers 27 cM (Supplemental Fig. 2B). Hence, although different chromosomes are involved in sex determination in the two genera, the linkage group bearing the sex determination region in ninespine sticklebacks has some of the same recombination characteristics as the threespine stickleback Y chromosome.
The genomic positions of the major sex determination loci are different in threespine and ninespine sticklebacks, yet it is possible that the same molecular mechanisms determine this fundamental trait in both genera. For example, both genera may have inherited the same sex determination mechanism from a common ancestor, but the gene(s) underlying this mechanism may be located on different chromosomes due to different evolutionary translocations, as has occurred in salmonids [36]. Identification of the genes controlling sex determination on LG12 of ninespine sticklebacks and chromosome 19 of threespine sticklebacks will permit a direct test of this hypothesis.
Multiple phenotypic traits cluster on the sex chromosome
Several other phenotypic traits mapped to LG12 in ninespine sticklebacks, including jaw length, head length, orbit (eye) diameter, and pectoral fin length (Supplemental Fig. 1, Supplemental Tables 3 and 5). With the exception of pectoral fin length, the phenotypic means for each of these traits were larger in male than female fish. Sexual dimorphism in head size and other skeletal traits has previously been demonstrated for wild populations of Pungitius [37, 38], and for wild and lab-bred Gasterosteus [39–44]. Clustering of phenotypic traits on the sex chromosome could be due either to pleiotropic effects of a single sex-determining locus, or to multiple loci controlling sexually dimorphic traits that are physically and genetically linked on the sex chromosomes. When we repeated our analysis of sex-linked traits using residuals with the average effect of sex removed, we no longer detected significant QTL on LG12, suggesting that most LG12 QTL are detecting male-female differences rather than effects of alternative chromosomes within males or females. However, we did detect significant differences between the two female haplotypes on LG12 for lateral plate phenotypes using the transformed data (p<0.05, ANOVA with Tukey’s Multiple Comparison Test), consistent with our QTL mapping of this trait described above.
Linkage between the primary sex determination locus and genes with differential effects in males and females is thought to be a key feature that drives evolution of sex chromosomes, including the accumulation of inversions and sequence divergence that suppresses recombination between the sex determining locus and neighboring genes [45]. Furthermore, the linkage between sex determination and LG12 in ninespine sticklebacks and at least one other stickleback species (Gasterosteus wheatlandi, the black-spotted stickleback) suggests that this chromosome “might have an abundance of genes with differential fitness effects in males and females and thus be predisposed to becoming a sex chromosome” [18]. The distinct linkage groups that control sex determination in threespine and ninespine sticklebacks will provide an excellent system to compare mechanisms of both sex determination and sex chromosome evolution in closely related lineages.
Genetics of convergent evolution
Several genetic studies have demonstrated that the same genes likely underlie similar changes among different animal lineages. For example, in Drosophila, Ultrabithorax and Ovo/shavenbaby control similar changes among different species in leg and abdominal trichome patterns, respectively, and repeated changes at the yellow locus control similar wing pigmentation in different species [46–49]. Among vertebrates, evolution of similar pigmentation phenotypes resulting from changes in the Melanocortin 1 receptor (Mc1r) in mammals, birds, and reptiles (reviewed in [50]); in Oculocutaneous albinism 2 in multiple populations of cavefish [51]; and in Kitlg in both sticklebacks and humans [10] demonstrate that independent changes in the same gene can generate broadly similar phenotypes in multiple lineages. In contrast, other examples of convergent morphological evolution appear to depend on different genetic mechanisms. For instance, complementation crosses suggest that regressive eye loss in blind Mexican cavefish has occurred by different mechanisms in different cave populations [52, 53]. While variant alleles of Mc1r control pigmentation phenotypes in the beach mouse (Peromyscus polionotus) and rock pocket mouse (Chaetodipus intermedius), exceptions to this genetic trend are known for each species [54–56]. Likewise, different genes in different species of Drosophila control similar changes in abdominal pigmentation [57].
Because recent genetic studies in threespine sticklebacks show that similar chromosome regions control similar phenotypes in many different populations [4–6, 8, 10, 29], we recognized at the inception of this study that genetic mapping in ninespine sticklebacks might largely identify the same chromosome regions. However, for every trait we examined, we found that the major loci controlling skeletal traits and sex determination in ninespine sticklebacks mapped to different regions than the major loci controlling the corresponding traits in threespine sticklebacks. The convergent evolution of lateral plate and skin color changes in different threespine stickleback popultions has often taken place by repeated selection of ancient variants of the Eda and Kitlg genes, respectively [8, 10]. These variants are present at low levels in migratory marine populations, and were presumably introduced into new locations when marine ancestors colonized new lakes and streams. Perhaps recent evolution from standing variation within a single species of stickleback is more likely to involve the same genes in different populations, whereas convergent evolution between more distantly related genera may be more likely to arise from independent mutations. The current study suggests that ninespine sticklebacks provide an outstanding system to find additional genes responsible for morphological diversity in natural populations of vertebrates, and to compare the detailed genetic basis of convergent evolutionary change in long-separated lineages.
Supplementary Material
Acknowledgments
We thank Frank von Hippel and the Department of Biological Sciences at the University of Alaska Anchorage for field assistance and laboratory facilities; the Broad Institute for the initial release of the threespine stickleback genome assembly; Katie Peichel, Craig Miller, and three anonymous reviewers for comments on the manuscript; Frank Chan, Pam Colosimo, Jonathan Craft, Katie Ellis, Tiffani Jones, Melissa Marks, and Sydney Stringham for experimental assistance. This work was supported by the Helen Hay Whitney Foundation, a Burroughs Wellcome Career Award in the Biomedical Sciences, and NSF grant IOS-0744974 (MDS); an NSF Graduate Research Fellowship (BRS); the University of Utah Biology Undergraduate Research Program (ALM); NSF grants DEB-0211391 and DEB-0322818 (MAB and F. J. Rohlf); and NIH Center of Excellence in Genomic Science grant 1P50HG02568 (DMK). DMK is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental data include Supplemental Experimental Procedures, two figures, and five tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: The Belknap Press of Harvard University Press; 2002. [Google Scholar]
- 3.Peichel CL, Nereng K, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 5.Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. Public Library of Science - Biology. 2004;2:635–641. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci U S A. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 9.Kimmel CB, Ullmann B, Walker C, Wilson C, Currey M, Phillips PC, Bell MA, Postlethwait JH, Cresko WA. Evolution and development of facial bone morphology in threespine sticklebacks. Proc Natl Acad Sci U S A. 2005;102:5791–5796. doi: 10.1073/pnas.0408533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory Changes in Kit Ligand Expression and Parallel Evolution of Pigmentation in Sticklebacks and Humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole NJ, Tanaka M, Prescott A, Tickle CA. Expression of limb initiation genes and clues to the basis of morphological diversification in threespine sticklebacks. Current Biology. 2003;13:R951–R952. doi: 10.1016/j.cub.2003.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Coyle SM, Huntingford FA, Peichel CL. Parallel evolution of Pitx1 underlies pelvic reduction in Scottish three-spined stickleback (Gasterosteus aculeatus) Journal of Heredity. 2007;98:581–586. doi: 10.1093/jhered/esm066. [DOI] [PubMed] [Google Scholar]
- 13.Bell MA, Stewart JD, Park PJ. The world’s oldest fossil threespine stickleback fish. Copeia (in press) [Google Scholar]
- 14.Nelson JS. Absence of the pelvic complex in ninespine sticklebacks, Pungitius pungitius, collected in Ireland and Wood Buffalo National Park Region, Canada, with notes on meristic variation. Copeia. 1971;1971:707–717. [Google Scholar]
- 15.Bell MA, Orti G, Walker JA, Koenings JP. Evolution of pelvic reduction in threespine stickleback fish: a test of competing hypotheses. Evolution. 1993;47:906–914. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen TR, Reisman HM. A comparative chromosome study of the North American species of sticklebacks (Teleostei: Gasterosteidae) Cytogenetics. 1970;9:321–332. doi: 10.1159/000130102. [DOI] [PubMed] [Google Scholar]
- 17.Ocalewicz K, opp-Bayat DF, Woznicki P, Jankun M. Heteromorphic sex chromosomes in the ninespine stickleback Pungitius pungitius. Journal of Fish Biology. 2008;73:456–462. [Google Scholar]
- 18.Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt G, Bell MA, Travis MP. Evolution toward a new adaptive optimum: phenotypic evolution in a fossil stickleback lineage. Evolution. 2008;62:700–710. doi: 10.1111/j.1558-5646.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 20.Blouw DM, Boyd GJ. Inheritance of reduction, loss, and asymmetry of the pelvis in Pungitius pungitius (ninespine stickleback) Heredity. 1992;68:33–42. [Google Scholar]
- 21.Giles N. The possible role of environmental calcium levels during the evolution of phenotypic diversity on Outer Hebridean populations of the three-spined stickleback, Gasterosteus aculeatus. Journal of Zoology (London) 1983;4:535–544. [Google Scholar]
- 22.Reimchen TE. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus: an adaptation to predators? Canadian Journal of Zoology. 1980;58:1232–1244. [Google Scholar]
- 23.Ziuganov VV, Zotin AA. Pelvic girdle polymorphism and reproductive barriers in the ninespine stickleback Pungitius pungitius (L.) from northwest Russia. Behaviour. 1995;132:1095–1105. [Google Scholar]
- 24.Zyuganov VV, Rosanov AS. Genetics of pelvic girdle reduction in fish (as illustrated by the ninespine stickleback Pungitius pungitius L.) Doklady Akademii Nauk SSR. 1987;293:155–159. [Google Scholar]
- 25.Shapiro MD, Marks ME, Peichel CL, Nereng K, Blackman BK, Jonsson B, Schluter D, Kingsley DM. Corrigendum: Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2006;7079:1014. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 26.Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisúa-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logan M, Tabin CJ. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 28.Shang J, Luo Y, Clayton DA. Backfoot is a novel homeobox gene expressed in the mesenchyme of developing hind limb. Dev Dyn. 1997;209:242–253. doi: 10.1002/(SICI)1097-0177(199706)209:2<242::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 2006;103:13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson JS. Comparison of the pectoral and pelvic skeletons and of some other bones and their phylogenetic implications in the Aulorhynchidae and Gasterosteidae (Pisces) Journal of the Fisheries Research Board of Canada. 1971;28:427–442. [Google Scholar]
- 31.Bell MA, Khalef V, Travis MP. Directional asymmetry of pelvic vestiges in threespine stickleback. J Exp Zoolog B Mol Dev Evol. 2007;308:189–199. doi: 10.1002/jez.b.21132. [DOI] [PubMed] [Google Scholar]
- 32.Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx1 and Pitx2 are required for development of hindlimb buds. Development. 2003;130:45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- 33.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 34.Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- 35.Mank JE, Promislow DE, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biological Journal of the Linnean Society. 2006;87:83–93. [Google Scholar]
- 36.Woram RA, Gharbi K, Sakamoto T, Hoyheim B, Holm LE, Naish K, McGowan C, Ferguson MM, Phillips RB, Stein J, et al. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chae BS, Yang HJ. Sexual dimorphism in eightspine stickleback, Pungitius sinensis: Gasterosteidae. Korean Journal of Zoology. 1990;33:260–265. [Google Scholar]
- 38.Kobayashi H. Cross-experiments with three species of stickleback, Pungitius pungitius (L.), Pungitius tymensis (Nikolsk), and Pungitius sinensis (Guichenot), with special reference to their systematic relationship. Journal of Hokkaido Gakugei University, Section B. 1959;10:363–384. [Google Scholar]
- 39.Kitano J, Mori S, Peichel CL. Sexual dimorphism in the external morphology of the threespine stickleback (Gasterosteus aculeatus) Copeia. 2007;2007:336–349. [Google Scholar]
- 40.Caldecutt WJ, Adams DC. Morphometrics of trophic osteology in the threespine stickleback, Gasterosteus aculeatus. Copeia. 1998;1998:827–838. [Google Scholar]
- 41.Caldecutt WJ, Bell MA, Buckland-Nicks JA. Sexual dimorphism and geographic variation in dentition of threespine stickleback, Gasterosteus aculeatus. Copeia. 2001;2001:936–944. [Google Scholar]
- 42.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): evidence for a species-pair in Paxton Lake, Texada Island, British Columbia. Canadian Journal of Zoology. 1992;70:361–369. [Google Scholar]
- 43.Albert AY, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- 44.Aguirre WE, Ellis KE, Kusenda M, Bell MA. Phenotypic variation and sexual dimorphism in anadromous threespine stickleback: implications for postglacial adaptive radiation. Biological Journal of the Linnean Society. 2008;95:465–478. [Google Scholar]
- 45.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 46.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 47.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 48.Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 49.Davis GK, Srinivasan DG, Wittkopp PJ, Stern DL. The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster. Dev Biol. 2007;308:621–631. doi: 10.1016/j.ydbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- 51.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkens H. Genetic interpretation of regressive evolutionary process: Studies of hybrid eyes of two Astyanax cave populations (Characidae, Pisces) Evolution. 1971;25:530–544. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 53.Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 54.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 55.Steiner CC, Rompler H, Boettger LM, Schoneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol Biol Evol. 2009;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- 56.Hoekstra HE, Nachman MW. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol Ecol. 2003;12:1185–1194. doi: 10.1046/j.1365-294x.2003.01788.x. [DOI] [PubMed] [Google Scholar]
- 57.Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc Natl Acad Sci U S A. 2003;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Ooijen J. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity. 1999;83:613–624. doi: 10.1038/sj.hdy.6886230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.