EMBO J 28, 2461–2468 (2009); published online 19 August 2009
DNA double-stranded breaks lead to the recruitment of the Rnf8–Ubc13 complex to the damage site where they catalyse K63-linked ubiquitination. Receptor-associated protein 80 (Rap80) subsequently binds the K63-linked chains and brings with it a BRCA1 complex. Sato et al (2009) solve the crystal structure of the Rap80-UIM1-UIM2:K63-linked diubiquitin complex to show that a contiguous helix spans the UIM1-UIM2 region and defines the relative orientation of the two UIMs for optimal binding to K63-linked chains. This study provides important insights into how protein ubiquitination triggers specific downstream signalling events.
Cells mount a multi-faceted response to genotoxic damage that involves transcriptional control, DNA repair machinery, and cell-cycle regulation. The arrangement of these events relies on post-translational modifications that expand the functionality of the target protein or control its cellular abundance. Ubiquitination is used for both of these purposes, and the mechanistic details of its many essential functions in DNA damage response are under intense scrutiny.
Substrates can be monoubiquitinated or polyubiquitinated with multiple ubiquitins attached directly to the substrate or serially to previously attached ubiquitin, thereby building a chain. Ubiquitin has seven lysines, creating opportunities for chain length and linkage diversity. DNA damage response uses multiple ubiquitin signalling mechanisms to communicate downstream events. K48-linked chains are generated on proteins involved in damage-induced cell-cycle arrest to promote their degradation and, thereby, limit the duration of their efficacy; this process controls Chk1 (Zhang et al, 2005), which is essential for the G2/M DNA damage-induced checkpoint. By contrast, monoubiquitination of proliferating cell nuclear antigen (PCNA) follows replication stalling at DNA damage sites and promotes the replacement of replicative DNA polymerases for damage-tolerant ones (for review, see (Ulrich, 2009).
The E3 ligase, Rnf8, and its associated E2 conjugating enzyme, Ubc13, are recruited to DNA double-stranded breaks, where they ubiquitinate the histone H2A variant, H2AX, with K63-linked chains (for review see (Yan and Jetten, 2008)). Rap80 joins the damage site through its affinity for K63-linked chains and brings with it a breast cancer suppressor protein (BRCA1) complex (Sobhian et al, 2007).
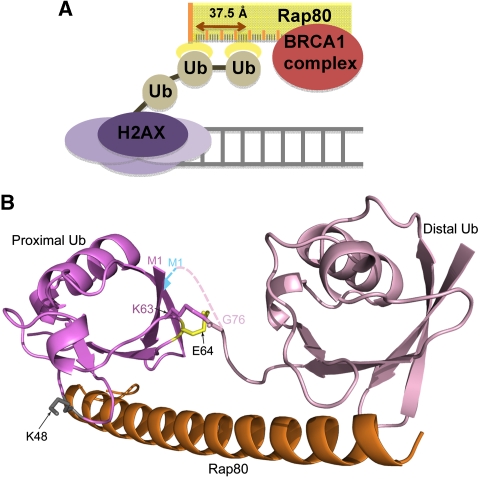
How does Rap80 specifically target K63-linked H2AX and not monoubiquitinated PCNA or K48-linked Chk1? It has two ubiquitin-interacting motifs (UIMs) and recently Sims and Cohen presented the surprising finding that Rap80's specificity for K63-linked chains is defined by the region between the two UIMs, and not the UIMs themselves (Sims and Cohen, 2009). In this issue of The EMBO Journal, Sato et al. present the crystal structure of Rap80-UIM1-UIM2 complexed with K63-linked diubiquitin to show that it forms a 60-Å long contiguous helix that restricts the relative orientation of the two UIMs to one optimized for binding to neighbouring ubiquitin subunits of K63-linked chains. Importantly, no contacts were observed between either of the UIMs and region linking the two ubiquitin subunits. This finding contrasts that of other ubiquitin receptors, in which one ubiquitin-binding domain simultaneously contacts two ubiquitin subunits and/or their linker regions to achieve specificity, such as hHR23a's C-terminal UBA domain (Varadan et al, 2005) or NEMO's UBAN motif (Rahighi et al, 2009). Hence, two UIMs are present in Rap80, not only to increase affinity, but also to define specificity through the intervening amino acids; these form a molecular ruler that directs the two ubiquitin-binding surfaces in roughly the same direction and separates their centres by 37.5 Å, two attributes optimal for binding to K63-linked chains (Figure 1A).
Figure 1.
Rap80 uses a molecular ruler mechanism to respond to K63-linked ubiquitin chains. (A) K63-linked ubiquitin chains at DNA damage sites attract Rap80, which brings a BRCA1 complex. Rap80's inter-UIM region acts as a molecular ruler by restricting its UIMs to a configuration optimized for binding to K63-linked ubiquitin chains. (B) Structure, solved by Sato et al., of Rap80-UIM1-UIM2:K63-linked diubiquitin is shown highlighting K48 (in grey) and the required changes for tandem binding with the M1 linkage (blue). E64 (shown in yellow) must rotate in M1-linked chains (data not shown) to avoid steric clashes with the linker region.
By contrast, the restrictions imposed by the inter-UIM region prohibit Rap80's UIMs from binding to ubiquitin subunits of K48-linked chains simultaneously, as these cannot adopt the required orientation (Figure 1B). This finding explains why Rap80 is not recruited to substrates conjugated with K48-linked chains. M1 and K63 are only 6.7 Å apart. Rap80 detects this spatial difference because linkage through M1 to form ‘linear chains' changes the permitted orientations of neighbouring ubiquitin subunits. When M1-linked diubiquitin is forced in silico into the configuration required for tandem binding to Rap80's UIMs, the proximal subunit's N-terminal β-strand is disrupted and its E64 adopts unfavourable backbone torsion angles to avoid steric clashes with a highly constrained diubiquitin linker region (Figure 1B).
Ubiquitin communicates with over 100 receptors for diverse outcomes. Sato et al. showed that ubiquitin-binding modules can serve as building blocks for larger structural entities to increase signalling stringency. This strategy parallels and complements the use of ubiquitin as an element for longer chains to achieve specificity. Yet, it is still remarkable that ubiquitination can explicitly command one receptor to a substrate. As Sato et al show Epsin1, which is involved in clathrin-mediated endocytosis, seems to apply the same strategy as Rap80 to confer preference for K63-linked substrates. So what prevents Epsin1 from being recruited to DNA damage sites and thus interfering with Rap80 signaling? No doubt the larger context also has a function in the communication between ubiquitinated substrates and their receptors. In this case, perhaps the BRCA1 complex is not passively shuttled by Rap80 but rather, actively participates in its localization to damage sites.
Acknowledgments
Research in KJW laboratory is supported by the National Institutes of Health (CA097004 and CA117888) and the American Cancer Society (RSG-07-186-01-GMC).
Footnotes
The authors declare that they have no conflict of interest.
References
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD (2009) Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 8: 461–469 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D (2005) Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell 18: 687–698 [DOI] [PubMed] [Google Scholar]
- Yan J, Jetten AM (2008) RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett 271: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin–proteasome pathway. Mol Cell 19: 607–618 [DOI] [PubMed] [Google Scholar]