Figure 5. Formation of some secondary structure depends on divalent ions and is important for the tertiary collapse of the Sc. ai5γ D135 ribozyme.
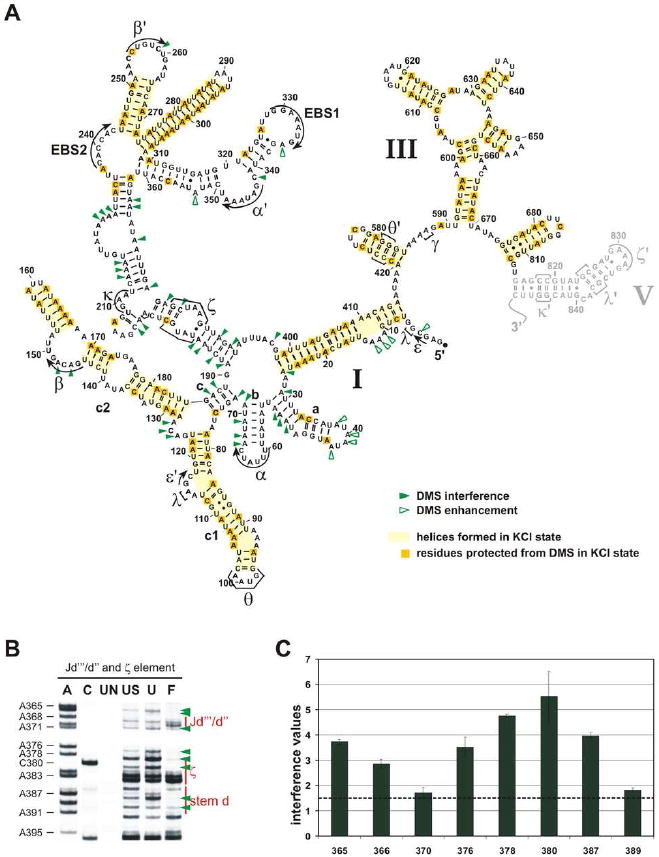
(a) The ‘KCl state’ and a summary map of observed DMS interferences and enhancements. Helical structures formed in the absence of any divalent ions (unfolded state) are highlighted in yellow (darker yellow boxes indicate the A and C residues that were not methylated by DMS). Green arrowheads mark the observed DMS interferences (closed symbol) or enhancements (open symbol). D5 is marked in gray, because it was not included in the analysis (see Methods). (b) Representative gels for the D5 docking site (ζ element) and flanking regions. The abbreviation scheme (UN, US, U, F) was explained in the legend to Figure 3; A and C refer to the sequence lanes which are required to identify the sites of interference. (c) Bar diagram shows the average value and standard deviation for each interference and enhancement found within the region shown in (b). The dotted line indicates the cutoff below which interferences and enhancements were considered insignificant.
