Abstract
Aims
In this article a review is made of data recently obtained on the structural diversity and possible functions of MADS box genes in the determination of flower structure in the African oil palm (Elaeis guineensis). MADS box genes play a dominant role in the ABC model established to explain how floral organ identity is determined in model dicotyledon species such as Arabidopsis thaliana and Antirrhinum majus. In the monocotyledons, although there appears to be a broad general conservation of ABC gene functions, the model itself needs to be adapted in some cases, notably for certain species which produce flowers with sepals and petals of similar appearance. For the moment, ABC genes remain unstudied in a number of key monocot clades, so only a partial picture is available for the Liliopsida as a whole. The aim of this article is to summarize data recently obtained for the African oil palm Elaeis guineensis, a member of the family Arecaceae (Arecales), and to discuss their significance with respect to knowledge gained from other Angiosperm groups, particularly within the monocotyledons.
Scope
The essential details of reproductive development in oil palm are discussed and an overview is provided of the structural and functional characterization of MADS box genes likely to play a homeotic role in flower development in this species.
Conclusions
The structural and functional data provide evidence for a general conservation of the generic ‘ABC’ model in oil palm, rather than the ‘modified ABC model’ proposed for some other monocot species which produce homochlamydeous flowers (i.e. with morphologically similar organs in both perianth whorls), such as members of the Liliales. Our oil palm data therefore follow a similar pattern to those obtained for other Commelinid species in the orders Commelinales and Poales. The significance of these findings is discussed.
Key words: Palm, MADS box, flower, Elaeis, monoecious, homeotic
INTRODUCTION: WHY STUDY FLOWER FORM?
Given the remarkable diversity of form displayed by flowers and their great utility as a morphological character in taxonomic studies, it is not surprising that they have been a focus of attention in recent years in evolutionary developmental biology. Innovations in flower structure were probably a key factor which contributed to the success of the angiosperms and the great diversification of lineages which occurred early in the evolution of this group is reflected in abundance of floral forms observed today in extant species. During the last decade, the development of molecular phylogenies has allowed the elucidation of phylogenetic relationships between most major angiosperm clades (Savolainen and Chase, 2003; Davies et al., 2004). Moreover, for an increasing number of angiosperm species, whole genome sequences are available, along with associated functional tools for molecular genetic studies. This has made it possible for researchers to carry out in-depth analyses of the molecular determination of flower structure in model plants such as thale cress (Arabidopsis thaliana), snapdragon (Antirrhinum majus) and rice (Oryza sativa) (Theissen et al., 2000). The large body of data thus obtained provides a useful starting point for studies in other higher plant taxa for which fewer molecular resources and functional tools are available.
In this article, we summarize recent work carried out on the structure and function of floral MADS box genes in the African oil palm, Elaeis guineensis (Arecoideae, Cocoseae; Dransfield et al., 2005), an economically important member of the palm family (Arecaceae), which constitutes the order Arecales within the monocotyledons. We compare and contrast our results with those obtained from other angiosperm lineages, especially within the monocotyledons. Gene structure/function studies on palms pose a number of technical difficulties, but are vital in order to understand how flower structure determination in the Arecaceae fits in with that of other groups.
FLOWERING IN OIL PALM
Flowering in Arecaceae
Palms are probably one of the most easily recognizable plant families, despite the relatively large size of the group – around 2400 species, according to a recent checklist (Govaerts and Dransfield, 2005). As noted by Tomlinson (1990), palms are characterized by generally highly branched inflorescences with a basal prophyll, a tendency towards monoecy and dioecy and the association of flowers within small groups (usually condensed cincinni) which are often characteristic of specific clades. A wide range of studies of inflorescence and flower development has been reported for palms (Tomlinson and Moore, 1968; Uhl, 1976; Uhl and Moore, 1977, 1978, 1980; De Mason et al., 1982; Uhl and Dransfield, 1984; Uhl, 1988; Barfod and Uhl, 2001; Stauffer et al., 2002; Rudall et al., 2003). As is typical of monocotyledonous species, palm species usually produce trimerous flowers. Some features have been noted as characteristic of specific groups, including the apocarpous character typical of (but not universal to) Coryphoid palms and the presence of distinct overlapping scales around the ovules of Calamoid species. Probably the most striking variation in flower structure is exhibited by the tribe Phytelepheae. This group is characterized by flowers with more than three organs per whorl and stamens which develop centrifugally in large numbers (from 120 to over 900 per flower; Uhl and Moore, 1977). With regard to their perianth, palm flowers often display distinguishable sepals and petals (Dransfield and Uhl, 1998). However, it is also common to observe a perigon-type perianth composed of two whorls of organs of similar appearance referred to as tepals. Floral bauplan forms part of the wide body of morphological and anatomical data which facilitate the classification of palm species into specific clades, complemented more recently by in-depth studies of molecular phylogeny (Dransfield et al., 2005). In this article, we focus our attention mostly on oil palm flower development as compared with other angiosperm families, particularly within the Liliopsida, for which an increasing body of molecular data is becoming available.
Reproductive development in oil palm
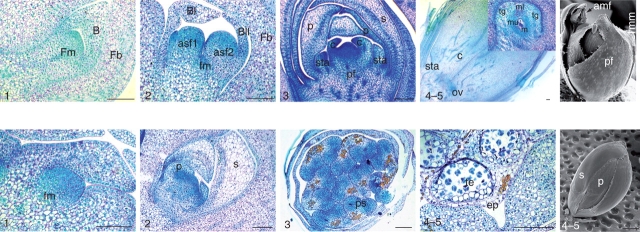
An in-depth microscopic analysis of oil palm inflorescence and floral development was carried out recently (Adam et al., 2005), complementing partial studies reported previously (Beinaert, 1935; Corley and Gray, 1976; Van Heel et al., 1987). The key developmental stages of inflorescence and flower development in E. guineensis are illustrated in Fig. 1. Oil palm is a long-lived single stemmed palm which bears, like the majority of palm species, a single vegetative shoot apical meristem maintained throughout the lifetime of the plant. Under favourable climatic conditions, this meristem is continuously active, producing a new leaf primordium approximately every 2 weeks in mature palms (Corley and Gray, 1976). The leaf takes 2–3 years to develop from initiation to the time when leaflets unfold in the centre of the palm crown. Inflorescences are formed throughout the year in the axils of their subtending leaves. Elaeis guineensis is monoecious, producing separate male and female inflorescences on the same palm in alternation, although mixed sex inflorescences are occasionally observed. Whereas the male inflorescence bears individual staminate flowers, the female inflorescence produces floral triads consisting of a pistillate flower flanked by two accompanying staminate flowers. The latter develop up to, and including, the appearance of microsporocytes in the pollen sac, after which no further development occurs and abscission takes place before the pistillate flower reaches maturity. In the perianth of oil palm flowers, sepals and petals are of a similar petaloid appearance, particularly in the pistillate flower. The reproductive organs of staminate flowers are composed of six stamens with connate filaments surrounding a pistillode, whereas pistillate flowers display rudimentary stamens (staminodes) and a gynoecium of three carpels.
Fig. 1.
Key stages of pistillate (upper panel) and staminate (lower panel) flower development in oil palm. Developmental stages (indicated at bottom left of each photo) were assigned on the basis of the differentiation of the floral whorls. Stage 1 corresponds to a floral meristem. Stage 2 corresponds to the initiation of perianth organs. Stage 3 corresponds to the development of perianth organs and the initiation of reproductive organs. Stage 4 corresponds to the development of reproductive organs and stage 5 to a mature flower. Photographs are of either PAS/NBR-stained transverse and longitudinal sections or scanning electron micrographs (right-hand photographs, upper and lower panels). Abbreviations: asf1/asf2, accompanying staminate flowers 1 and 2; B, bracteole; BI/BII, bracteoles I and II; c, carpel; ff, pistillate flower; fm, floral triad meristem; Fb, floral triad bract; o, ovule; p, petal; s, sepal; sta, staminodes; c, carpel; m, megaspore mother cell; ps, pollen sac; sta, staminodes; te, tetrads; tg, integuments.
A homeotic floral variant in oil palm: the mantled abnormality
For the purpose of understanding the molecular processes which determine flower structure in oil palm, a previously described homeotic epimutant, known as mantled (Corley et al., 1986) is of particular interest. Mantled palms exhibit a transformation of stamens and staminodes into carpel-like structures in staminate and pistillate flowers, respectively (Adam et al., 2005). In the mantled staminate flower, the transformation of the stamens into pseudocarpels results in sterility, whereas in the mantled pistillate flower, fertilization may occur in less severe cases to produce characteristic fertile fruits. In more severe cases, parthenocarpy or arrested development occurs. The mantled phenotype is observed in oil palms regenerated from tissue culture (Corley et al., 1986) and may be transmitted through meiosis (Rao and Donough, 1990). However, reversion to wild type is observed in the field in some but not all individuals (Durand-Gasselin et al., 1990), indicating an epigenetic origin. In the female inflorescence of mantled palms, all stages up to and including reproductive organ initiation appear the same as in normal palms. Developmental divergence occurs shortly afterwards, when organs resembling carpels are seen to develop in the androecium in place of staminodes. These carpeloid structures lack ovules and are thus sterile. In the case of the mantled staminate flower, the divergent developmental pattern is witnessed at the same stage, i.e. during the elongation of the organs of the third whorl, which are seen to display a central vascularization characteristic of carpels, whereas stamens normally have a peripheral vascularization (Fig. 1). The homeotic transformation of stamens to sterile carpel-like structures may also be observed in the accompanying staminate flowers of floral triads on the female inflorescence. As previously demonstrated with model flowering plants, the study of floral variants in oil palm is likely to provide a useful means to understand the molecular mechanisms which regulate floral morphology in this species.
THE MOLECULAR BASIS OF FLOWER STRUCTURE: WHAT DO WE KNOW FROM OTHER SPECIES?
The ABC model
Genetic studies performed on model species, including Arabidopsis thaliana and Antirrhinum majus, led in the early 1990s to the identification of regulatory pathways and genes which control various aspects of flowering in a wide range of plants (Coen and Meyerowitz, 1991; Levy and Dean, 1998; Meyerowitz, 1998). At the outset, molecular genetic studies of higher plant flowering revealed the existence of a generalized floral signalling hierarchy within which individual genes were found to act at specific levels. Several classes of genes were identified in this way, including those determining flowering transition, inflorescence meristem identity and floral organ identity (Okada and Shimura, 1994; Weigel, 1995). An important advance was achieved in the formulation of the ABC model to explain floral organ identity determination based on studies performed on A. thaliana and Antirrhinum (Coen and Meyerowitz, 1991). According to this model, the identity of each whorl of the flower is governed by the expression of one or more homeotic genes of function A, B or C. Expression of the A-class function alone specifies sepal formation. The combination of A- and B-class functions specifies the development of petals, and the combination of B- and C-class functions results in the formation of stamens. The expression of the C function alone determines the development of carpels. Since its initial conception, this model has been modified to take account of newer data, revealing a D-type activity involved in the specification of ovules (Angenent and Colombo, 1996) and an E function necessary for the determination of the corolla, androecium and gynoecium (Pelaz et al., 2000). The essential details of the current generic ABC model are shown in Fig. 2 (top right).
Fig. 2.
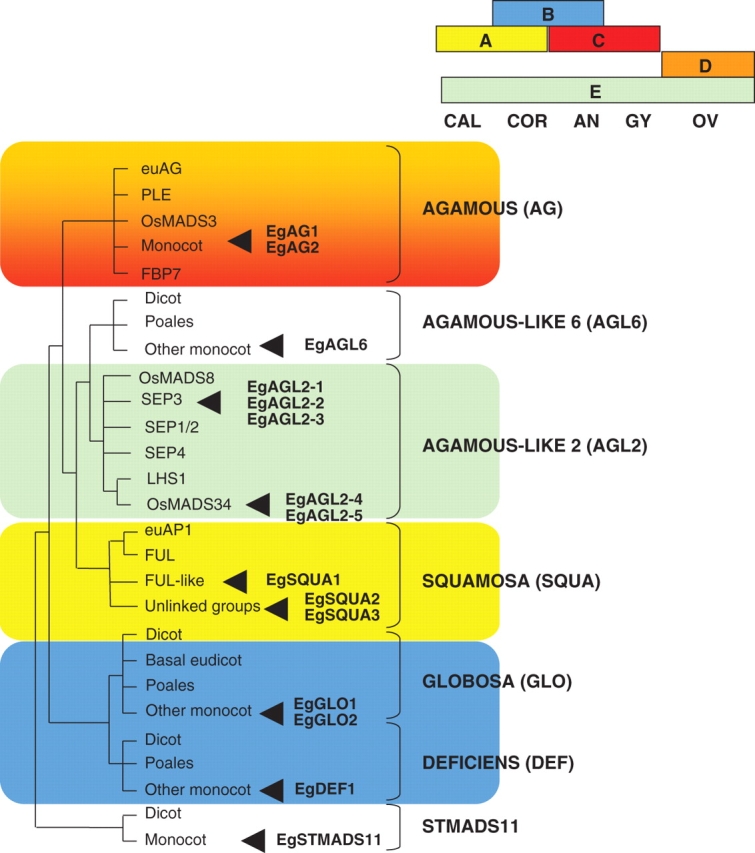
Dendrogram illustrating sequence affinities between oil palm MADS box proteins and selected relatives from other angiosperm groups. Subfamilies are designated according to the system of Becker and Theissen (2003). The tree shown is a schematic representation of topologies obtained using the maximum parsimony (MP) method with full-length MADS box amino acid sequences. Abbreviations: euAG, euAGAMOUS clade; PLE, PLENA clade; FBP7, FLORAL BINDING PROTEIN 7 clade; SEP3, SEPALLATA3; SEP1/2, SEPALLATA1/2; LHS1, LEAFY HULL STERILE1; euAP1, euAPETALA1 clade; FUL, FRUITFULL; CAL, calyx; COR, corolla; AN, androecium; GY, gynoecium; OV, ovule.
Despite this overall conservation of gene function between species, several limitations to the generic organ identity model have been identified, including the apparently poor conservation of A-type function, which is likely to be a relatively recent development in higher plant evolution (Egea Gutierrez-Cortines and Davies, 2000). Another aspect of flower structure determination which appears increasingly more complex in reality than in the ABC model is the distinction of roles between C and D genes, which in A. thaliana are closely related and overlap in their functions (Favaro et al., 2003; Pinyopich et al., 2003). The frequent occurrence of paralogues in lineages involved in flower structure determination makes gene structure/function studies complicated; however, the duplications from which they have arisen will in some cases have been important evolutionary events.
The key role of MADS box genes in flower development
Nearly all floral homeotic genes, including those mentioned above, code for MADS box transcription factors. These proteins are common to all eukaryotic groups; however those found in higher plants are distinguishable by their characteristic MIKC structure, referring to the four different domains which they possess (Theissen et al., 2000). Phylogeny reconstructions reveal that the MADS box gene family is composed of a number of defined gene clades (Becker and Theissen, 2003). In eudicotyledons, 14 different paralogous MIKC-type MADS box gene subfamilies, have been defined (Alvarez-Buylla et al., 2000; Becker et al., 2000; Theissen et al., 2000, Becker and Theissen, 2003). Thanks to studies performed on a number of different species, strong structure/function relationships have been established for a number of these groups. Thus A function has been inferred for certain members of the SQUAMOSA (SQUA) subfamily, B function for genes of the GLOBOSA (GLO) and DEFICIENS (DEF) classes, C and D functions for members of the AGAMOUS (AG) group and E function for various AGAMOUS-like2 (AGL2) genes. We took advantage of these conserved structure/function relationships in order to study genes likely to regulate flower structure in oil palm.
MADS BOX GENES IN OIL PALM
In our studies, MADS box genes of oil palm were identified and characterized via the isolation of full-length cDNAs. Most sequences were isolated by PCR amplification with degenerate MADS box-specific primers using cDNA derived from male or female inflorescences. The resulting PCR fragments were then used to screen cDNA libraries prepared from the same type of plant material (Adam et al., 2006). An additional MADS box gene was identified by systematic sequencing of cDNA clones as part of an EST (expressed sequence tag) collection (Jouannic et al., 2005). As a result, 15 different oil palm MADS box genes were identified and named according to their sequence affinities as follows: EgSQUA1, EgSQUA2 and EgSQUA3 (SQUAMOSA or SQUA group); EgDEF1 (DEFICIENS or DEF group); EgGLO1 and EgGLO2 (GLOBOSA or GLO group); EgAG1 and EgAG2 (AGAMOUS or AG group); EgAGL2-1, EgAGL2-2, EgAGL2-3, EgAGL2-4 and EgAGL2-5 (AGAMOUS-like2 or AGL2 group); EgAGL6-1 (AGAMOUS-like6 or AGL6 group); and EgSTMADS11-1 (STMADS11 group). Genes were assigned initially to MADS box subfamilies on the basis of sequence similarities. The overall sequence relationships of the proteins encoded by the oil palm MADS box genes are shown pictorially in the dendrogram in Fig. 2, which also reveals the different clades identified within each subfamily. These data were described in detail previously (Adam et al., 2006) except for the AGL6 and STMADS11 groups. For this study, entire amino acid sequences were used in conjunction with the maximum parsimony (MP) method. A summary of essential details of the oil palm MADS box genes and the proteins which they encode is given in Table 1. This Table includes information on the possible existence of closely related genes as evaluated by Southern hybridization and their specificity of expression as revealed by RT-PCR analysis on several different organs/developmental stages of the plant.
Table 1.
Summary of data obtained on the complexity of the different oil palm MADS box groups, on the spatio-temporal expression patterns of five selected oil palm MADS box genes and on the phenotypic effects induced when they were overexpressed in transgenic Arabidopsis thaliana plants
| Gene | Size of cross-hybridizing gene group | Expression pattern |
Phenotype when ectopically expressed in A. thaliana | Possible role(s) | ||
|---|---|---|---|---|---|---|
| Staminate flowers | Pistillate flowers | Mantled palms | ||||
| EgSQUA1 | 2 | Inflorescence and floral meristems | Inflorescence and floral meristems | Unaffected | Tall phenotype (larger number of nodes) | Inflorescence/ floral meristem identity (A function?) |
| EgDEF1 | 1 | Stamens and petals | Staminodes and petals | Decreased expression (both sexes) | No alterations observed | B function |
| EgGLO2 | 2 | Sepals, petals and stamens | Sepals and petals | Decreased expression (both sexes) | Transformation of sepals to petals | B function |
| EgAG2 | 2 | All whorls (at immaturity) | Carpel primordia/ ovules | Decreased expression (mainly in pistillate flower) | No alterations observed | C and/or D function |
| EgAGL2-1 | 2–5 | Petals and stamens | Petals and ovule primordia | Decreased expression (both sexes) | Leaf-Like sepals and petals; ‘flower within a flower’ | E function |
In subsequent work, we focused our attention on those groups for which a role in floral organ identity determination has been demonstrated, namely the SQUA (A function), GLO (B function), DEF (B function), AG (C/D functions) and AGL2 (E function) subfamilies. As can be observed from the dendrogram in Fig. 2, most subfamilies can be further resolved into smaller clades, some specific to monocots or dicots. Some sequences were observed to be rooted in an unresolved fashion at the base of their subfamily group. This included in the case of oil palm the SQUA homologues EgSQUA2 and EgSQUA3. All other oil palm MADS box sequences were found to branch with related sequences from other species within their subfamily, providing some initial clues as to their possible functions.
POSSIBLE FUNCTIONS INFERRED FROM EXPRESSION ANALYSIS AND TRANSGENIC STUDIES
The phylogenetic reconstructions carried out for each MADS box subfamily provide a useful insight into evolutionary relationships both within the group of oil palm genes studied and also with respect to other plant taxa. Although this may provide clues as to the possible roles of the oil palm genes, studies of a functional nature are essential in order to validate any hypotheses made. The investigation of floral gene function in oil palm is complicated by the large size and long life cycle of the plant, flowering occurring only about 3 years after germination. Thus, although genetic transformation of oil palm has been achieved (Parveez et al., 2000), transgenic studies of floral gene function in this species would require many years to yield results. In order to circumvent this problem, we employed ectopic expression in A. thaliana (under the control of the CaMV 35S promoter) as a means of assessing the conservation of MADS box protein function between oil palm and dicots. The data obtained by genetic transformation were coupled with information on the spatial and temporal expression patterns of each gene in normal and mantled palms, obtained by in situ hybridization and RT-PCR, respectively, so as to propose putative functions. Five of the oil palm MADS box genes were selected for detailed investigations of their possible function in oil palm reproductive development (Adam et al., 2007). On the basis of their DNA sequences, the genes selected showed similarities with various different homeotic genes implicated in the ABC model: EgSQUA1 (A class and/or meristem identity); EgGLO2 and EgDEF1 (B class); EgAG2 (C or D class); and EgAGL2-1 (E class). The isolation of MADS box cDNAs from oil palm was also recently described by Alwee et al. (2006), who identified genes similar or identical to each of the above, with the exception of EgDEF1.
Petal and sepal identities are clearly distinguished by MADS box gene expression in Elaeis guineensis
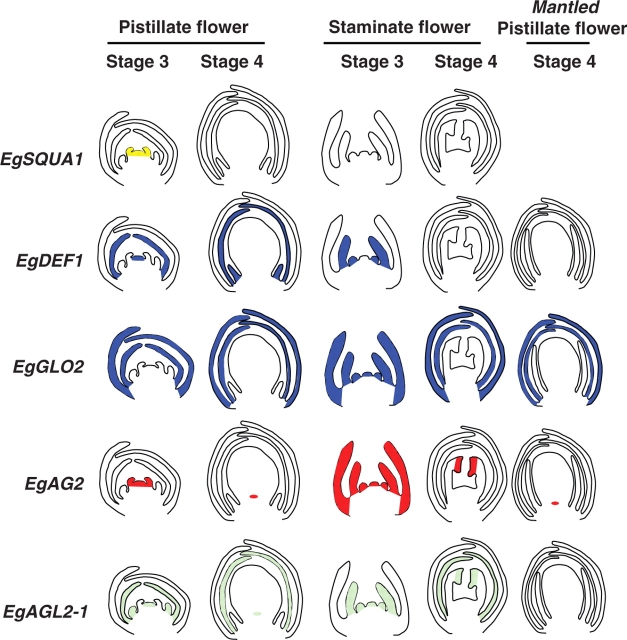
Within the general objective of identifying possible functions for floral MADS box genes in oil palm, we wished to address the specific question of whether petals and sepals have distinct identities in this species, and whether the mantled flower abnormality involves a change of organ identity in the perianth, as seen in the B-class mutants of model species. It is important to deal with this question at the outset when interpreting the data obtained, in order to establish the overall framework within which ABC functions can be attributed to specific genes. Elaeis guineensis produces, like many monocot species, flowers containing an inner and outer perianth of similar appearance, the petals and sepals sometimes being referred to collectively as tepals. In the case of the pistillate flower, as noted by Beinaert (1935), the only major difference between the inner and outer perianth whorls is in the size of the organs produced. Beinaert therefore considered the pistillate flower to be homochlamydeous (i.e. with morphologically similar organs in both perianth whorls). In the case of the staminate flower, some colour distinction exists between the inner and outer perianth organs, the former being more translucent in appearance. In the palm family as a whole, the morphological distinction of petals and sepals is possible for many species (Dransfield and Uhl, 1998). Our results indicate that the identities of petals and sepals can be distinguished by MADS box gene expression patterns in the case of oil palm. This is well illustrated by the EgDEF1 gene, for which expression was detected in the petals but not in the sepals of pistillate flowers (Fig. 3 and Table 1). EgDEF1 was also observed to be expressed in the androecium of flowers of both sexes. This spatial expression pattern is typical of classical B-class genes in model species such as A. thaliana in which the calyx and corolla are structurally different, as will be discussed below. Differences between sepals and petals of oil palm were also revealed by the spatial expression pattern of EgAGL2-1, a SEPALLATA3 (SEP3)-like gene of putative E function.
Fig. 3.
Spatial expression patterns of oil palm MADS box genes in staminate and pistillate flowers: schematic representation of in situ hybridization results. Two different developmental stages are depicted: firstly, the developing androecium and gynoecium stage (Stage 3); and secondly, the stage where gynoecium and androecium have reached full size but not maturity (Stage 4). Observations with mantled pistillate flowers are shown for Stage 4 only. Floral zones in which an in situ hybridization signal was observed are shown in colour, depending on the gene involved (yellow for EgSQUA1, blue for the B class genes EgDEF1 and EgGLO2, red for EgAG2 and green for EgAGL2-1).
Utility of the mantled epimutant for the elucidation of floral homeotic functions in oil palm
Floral mutants are an invaluable tool for studies aimed at elucidating gene function. In the case of oil palm, for which genetic approaches would require dauntingly long periods of time, we capitalized on the availability of the mantled epimutant induced by tissue culture. The mantled phenotype resembles that of B-class mutants, such as apetala3 (ap3) and pistillata (pi) of Arabidopsis thaliana (AP3 is a DEF-type gene and PI a GLO-type one) in which stamens are homeotically transformed to carpel-like structures. The ap3 and pi mutants also exhibit a conversion of petals into sepals. We therefore investigated whether a petal to sepal transformation could be revealed in mantled palms by MADS box gene expression changes. Our results corroborated this hypothesis; for example, no EgDEF1 transcripts were detected in the inner perianth of mantled flowers, implying that a homeotic conversion had occurred.
More generally, our RT-PCR results (summarized in Table 1) revealed that the expression of four of the five MADS box genes investigated was lower in mantled palms. This raises interesting questions as to the identity of the gene or genes initially perturbed in their activity in epigenetically altered plants. It seems unlikely that several different genes would all be directly perturbed; thus one hypothesis which can be made is that the initial gene target is an upstream regulator of the MADS box genes. This hypothesis suffers from the drawback that mutations in the genes which regulate organ identity genes generally produce much wider phenotypic effects than those seen in mantled palms. For example, mutations in the UNUSUAL FLORAL ORGANS (UFO) gene result in changes not only to floral organ identity, but also to inflorescence structure (Hepworth et al., 2006). To account for the specific nature of the mantled abnormality, it would therefore be necessary to invoke the existence of a target gene possessing either a novel function or exercising a much narrower role than its orthologues in previously described systems. An alternative and probably more plausible way to explain why several different MADS box genes show reduced expression in mantled plants is based on the fact that MADS box genes can regulate each other. Indeed many MADS box genes contain in their promoter regions regulatory sequences known as CArG boxes which are themselves binding sites for MADS box transcription factors. Binding may often occur to heteromultimeric complexes containing two or more different MADS proteins, thus the interactions taking place are complex. In the context of the mantled abnormality, it is interesting to note that in A. thaliana, mutations in the APETALA3 gene result in reduced transcript levels of the PISTILLATA gene and vice versa (Goto and Meyerowitz, 1994). Thus it is possible that one of the MADS box genes described here might be the initial genomic target of the epigenetic somaclonal variation event with secondary consequences on the expression of other MADS box genes. Further studies will be required to reveal the identity of the gene(s) involved and the exact nature of the epigenetic deregulation which has occurred.
Summary of functional data obtained: a model to explain the possible functions of five different floral MADS box genes of oil palm
Table 1 and Fig. 3 summarize the data obtained from gene expression and transgenic studies. For each of the genes investigated, a possible function is proposed on the basis of available results.
In the case of EgSQUA1, a role in the determination of inflorescence and floral organ identity is suggested. EgSQUA1 expression is concentrated in meristematic zones of the inflorescence and flower and a tall phenotype results when the gene is overexpressed in transgenic A. thaliana plants due to a larger number of nodes produced during the reproductive phase of the plant.
In the case of EgDEF1, its specific expression pattern in petals and sepals, coupled with the observed reduction in transcripts associated with the mantled abnormality, argue strongly in favour of a B function, as suggested by its sequence affinities.
In the case of EgGLO2, a functional similarity with the A. thaliana PISTILLATA gene was observed, since both produce the same phenotypic changes when ectopically expressed in transgenic plants of the latter species, namely the transformation of sepals into petals. Using the same approach, an identical phenotype was observed by Alwee et al. (2006) for the oil palm gene EgMADS16, a paralogue of EgGLO2 also isolated in our laboratory under the name of EgGLO1. In their study, Alwee et al. demonstrated that EgMADS16 was able to complement an A. thaliana pistillata mutant.
The homeotic modifications seen in 35S:EgGLO2 transgenic plants, taken together with the observed reduction of EgGLO2 transcripts in mantled palms, suggest a B function for this gene. Nevertheless, EgGLO2 appears to diverge compared with its A. thaliana relative inasmuch as transcripts of this gene are detected in sepals. This observation should be interpreted with caution, given that it does not necessarily signify the accumulation of the EgGLO2 protein. Indeed, in Lilium longiflorum, it has been shown that transcripts of the DEF gene LMADS1 accumulate in all whorls of the flower, but that the corresponding protein is present only in petals and stamens (Tzeng and Yang, 2001). An additional factor to bear in mind is that according to the generic ABC model, the GLO protein cannot exercise a B function on its own, but requires the presence of DEF, with which it forms a heterodimer and probably other types of heteromultimeric complexes (Theissen and Saedler, 2001). Nevertheless, it appears that in the Liliales at least, GLO-type proteins possess the capability of binding in vitro in a sequence-specific manner to DNA as a homodimer (Winter et al., 2002; Kanno et al., 2003).
On the basis of in situ hybridization studies, a C and/or D function can be proposed for EgAG2, since transcripts were detected both in ovule primordia and more generally in the carpel, as well as in other parts of the flower at earlier stages. The existence of genes possessing a mixed C/D function has already been demonstrated in A. thaliana (Favaro et al., 2003; Pinyopich et al., 2003).
In the case of EgAGL2, ectopic expression in transgenic A. thaliana produced an altered flower phenotype similar to the quadruple sepallata (sep1/sep2/sep3/sep4) mutant (Ditta et al., 2004). It was noted that sepals and petals acquired leaf-like characteristics; also a ‘flower within a flower’ was observed in place of the ovule in the gynoecium. This phenotype is hypothesized to be due to a dominant negative effect whereby the oil palm EgAGL2-1 protein is able to bind to the SEP3 protein partners in A. thaliana without possessing all necessary specificities for biological activity. Collectively, these data argue for an E function for EgAGL2. The AGL2-like subfamily appears to have expanded and functionally diversified in the monocots compared with dicots; nevertheless it is interesting to note that E function was recently demonstrated to be conserved in rice through studies of loss of function mutants of the OsMADS1 gene (Kumar et al., 2005).
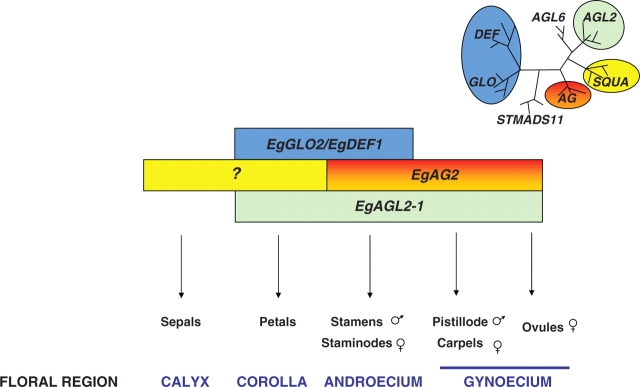
By combining the functional data, we were able to propose a tentative model to explain the action of the five oil palm MADS box genes studied in detail (Fig. 4). This model represents in essence the generic ABC model and distinguishes oil palm from some other monocot species, as is discussed below.
Fig. 4.
A model to explain the possible roles of various oil palm genes in the determination of flower structure in oil palm, as based upon the generic eudicot ABC model (Coen and Meyerowitz, 1991; Angenent and Colombo, 1996; Pelaz et al., 2000). In the top left-hand corner is shown a schematic representation of sequence relationships between the oil palm MADS box genes studied. Boxes are colour shaded according to homeotic functions as follows: yellow, A function; blue, B function; orange, C and D functions; green, E functions.
ABC model variation amongst the monocotyledons
Since the ABC model was first conceived for eudicots, a number of studies have been undertaken to investigate whether floral organ identity is determined in a similar way in monocots. Figure 5 provides a summary of current knowledge of floral organ identity regulation within the Liliopsida, with putative ABC models shown where sufficient data exists to allow hypotheses to be made.
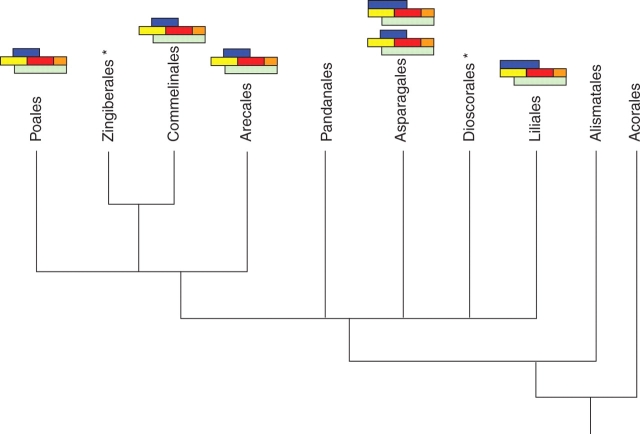
Fig. 5.
Summary of MADS box structural and functional data obtained for monocot orders. The characterization of species putatively following the generic ABCDE model or ‘modified ABC model’ (involving B function in the outer perianth) is indicated where data are available. Orders for which sequences are available but not functional, data are indicated by an asterisk. Note that the tree (not to scale) is for illustrative purposes only and is based on the topology indicated by Savolainen and Chase (2003), which is undergoing revision.
With regard to the determination of floral reproductive organs, studies on a wide range of species have revealed that there is a structural conservation of C and D MADS box gene lineages in monocots (Skipper et al., 2006). A general, if not universal conservation of C- and D-type gene expression patterns in monocots with respect to dicots suggests a corresponding functional conservation, and has been corroborated by transgenic studies in some cases (e.g. Kyozuka and Shimamoto, 2002; Tzeng et al., 2002; Benedito et al., 2004). More generally, global conservation of the generic ABC model has been demonstrated for some species. This was found to be the case for the Poales, the first monocot group to be studied in detail (Kramer and Jaramillo, 2005). B-class genes have been a matter of particular interest in monocot studies, since their activities appear to be important in the distinction of petals and sepals, which are sometimes similar in appearance in this group. Studies on members of the Poales such as maize and rice (Ambrose et al., 2000; Nagasawa et al., 2003) revealed that B-class gene expression followed a similar pattern to that seen in model eudicots, B-type activity being detected in lodicules but not in paleae/lemmae. Another monocot order in which the generic ABC model appears to be conserved is the Commelinales (Ochiai et al., 2004). In this case, two species were studied, namely Tradescantia reflexa and Commelina communis, both of which display distinct petal and sepal morphologies. In both plants, the expression of DEF and GLO genes was found to clearly mark petal identity compared with that of the sepals, although in the case of T. reflexa, a second GLO paralogue was identified and found to be expressed in sepals.
In contrast to the Poales and Commelinales, species from some other monocot clades were found not to conform to the generic ABC model. In the Liliales, studies of two species have been reported, namely tulip (Tulipa gesneriana; Kanno et al., 2003) and lily (Lilium longiflorum; Tzeng and Yang, 2001). In these two cases, a perianth containing similar petaloid organs in both whorls is produced. In order to explain how this floral configuration might be determined, an altered model was proposed whereby B-class gene expression is expanded to include the outer perianth. This was referred to as the ‘sliding boundary model’ (Kramer et al., 2003) or ‘modified ABC model’ (Van Tunen et al., 1993). In the former case, the model was proposed to take account of data obtained with species of the Ranunculaceae. The studies performed by Tzeng et al. (2001) and Kanno et al. (2003) provided support for the modified ABC model in Lilium longiflorum and Tulipa hybrida, respectively.
In contrast with the other monocot orders mentioned, data obtained on species of the Asparagales revealed divergences within the group, with Agapanthus praecox apparently conforming to the modified ABC model (Nakamura et al., 2005) and Asparagus officinalis not (Park et al., 2003, 2004). This is particularly surprising given that the flowers of A. officinalis display two whorls of almost identical petaloid organs in their perianth; thus it could be hypothesized that a divergent signalling pathway for petaloid tepal specification exists in this species. Nevertheless, as pointed out by Kramer and Jaramillo (2005), some caution should be exercised when comparing data between the different species, since the developmental stages studied were not always the same and in the case of L. longiflorum, only a single late stage was investigated.
Although the data available on ABC gene function in monocots are as yet fragmentary, one general trend which can be observed in Fig. 5 is the conservation of a molecular distinction (in the form of B gene expression) between the two perianth whorls within the Commelinid group, which includes palms. In the specific case of oil palm, in which this molecular distinction is not accompanied by clear morphological differences, it can be hypothesized that additional signalling mechanisms have evolved in organ identity determination, as suggested also for A. officinalis (Park et al., 2003, 2004). Outside the Commelinid group, the modified ABC model might well be a characteristic feature of the Liliales, but this appears not to be the case for the Asparagales, in which both types of ABC scenario can be found.
When comparing B gene data for the different monocot clades, it is interesting to note that the presence of multiple GLO (but not DEF) genes in many cases. In addition to the cases of E. guineensis (Arecales) and Tradescantia reflexa (Commelinales) mentioned previously, other examples include: the Burmese fishtail palm Caryota mitis (Arecales; Genbank accessions AAY56602 and 56603); rice, maize and wheat (Poales; Chung et al., 1995; Münster et al., 2001; Hama et al., 2004), Asparagus officinalis (Asparagales; Park et al., 2004), the Martagon lily Lilium martagon (Liliales; Genbank accessions AAY56593 and AAY56594) and the ornamental banana Musa ornata (Zingiberales; Genbank accessions AAY56605 and AAY56606). This contrasts with dicots, in which often only a single GLO gene is observed and suggests that a GLO gene duplication occurred early in the evolution of the monocots (Zahn et al., 2005). Such an event is likely to have been followed by functional diversification, which might account for the unexpected expression patterns of certain GLO genes in vegetative tissues of both monocots and dicots (Southerton et al., 1998; Münster et al., 2001; Skipper, 2002; Kanno et al., 2003). In the case of oil palm, EgGLO2 (but not EgGLO1) transcripts were detected in roots (Adam et al., 2006).
In summary, the very limited data available provide evidence for an overall conservation of the generic ABC model within the Commelinid clade. In contrast, the modified ABC model can be applied to species in the more phylogenetically distant Liliales, while in the case of the Asparagales, both types of scenario can be found. It would clearly be of great interest to know which of the two models described (if either) represents the plesiomorphic character of the monocots and indeed of the angiosperms. The resolution of this question is complicated by the great diversity of flower structure which exists amongst basal angiosperms and the fact that the calyx (and possibly even the corolla) is generally considered to have arisen independently several different times during evolution (Zanis et al., 2003). This question and others need to be addressed by wider and deeper sampling of molecular data amongst monocots, so as to obtain a better evolutionary perspective and to identify new gene orthologues and paralogues. It should be borne in mind that at least some floral bauplan diversity in monocots may not be attributable to the functioning of ABC MADS box genes. An interesting illustration of this point is the lodicule of the grass flower, which is considered to be a floral morphological innovation and which expresses B-class MADS box genes despite its non-petaloid characteristics (Kramer and Jaramillo, 2005). No doubt novel floral gene functions will also have arisen during the evolution of the palm family.
On a more general level, the sequence and functional data described here for oil palm should help to shed further light on the occurrence of duplication, neofunctionalization and subfunctionalization amongst MADS box genes during the evolution of the monocots. In the absence of a complete sequence of the oil palm genome, the current picture is inevitably fragmented; nevertheless, our data tend to confirm trends observed in other species. Apart from the example of GLO gene duplication mentioned above, another subfamily in which gene duplication and functional diversification is likely to be prevalent is the AGL2 group. Indeed, five different oil palm genes of this clade have been identified and found to display divergent expression patterns (Adam et al., 2006, 2007), a situation similar to that occurring in other monocots such as rice (Malcomber and Kellogg, 2005). Given that the expression of all five oil palm AGL2 genes characterized is specific to the inflorescence, their functions are likely to be restricted to the development of the reproductive structures of the plant, as is typical but not universal in this MADS box subfamily.
One aspect of reproductive development in oil palm upon which the current data do not shed light is sex determination, none of the oil palm MADS box genes described having been found to display a sex-dependent expression pattern. Given that there appears to be no common mechanism for sex determination in higher plants (Ainsworth, 2000), it is impossible to speculate on the nature of the genes involved in this process in oil palm, although it is interesting to note that sex-dependent expression of MADS box genes has been observed in some dioecious plants such as Rumex acetosa (Ainsworth et al., 1995).
CONCLUSIONS
MADS box genes probably lie at the heart of many key evolutionary events in plants through the fundamental role which they play in the regulation of reproductive development in general and floral structure in particular. Our data on the structure, expression and functional analysis of oil palm MADS box genes help to fill in a gap in existing knowledge and will allow the palm family to be compared and contrasted with other groups which have traditionally received more attention in this field. It is hoped that in future years, new information will be gathered from other members of the Arecaceae. This should help in the long term to provide an insight into the regulatory processes which underlie the rich structural diversity seen within the family.
ACKNOWLEDGEMENTS
We thank two anonymous referees for helpful comments in the improvement of the manuscript. We gratefully acknowledge the help of colleagues in Malaysia (FELDA Agricultural Services), Costa Rica (ASD, Coto) and Côte d'Ivoire (CNRA, La Mé Station) in the provision of plant material. This work was supported by an MESR PhD grant to H.A. and institutional funding from IRD and CIRAD.
LITERATURE CITED
- Adam H, Jouannic S, Escoute J, Duval Y, Verdeil JL, Tregear JW. Reproductive developmental complexity in the African oil palm (Elaeis guineensis) American Journal of Botany. 2005;92:1836–1852. doi: 10.3732/ajb.92.11.1836. [DOI] [PubMed] [Google Scholar]
- Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear JW. MADS box genes in oil palm (Elaeis guineensis): patterns in the evolution of the SQUAMOSA, DEFICIENS, GLOBOSA, AGAMOUS and SEPALLATA subfamilies. Journal of Molecular Evolution. 2006;62:15–31. doi: 10.1007/s00239-005-0333-7. [DOI] [PubMed] [Google Scholar]
- Adam H, Jouannic S, Morcillo F, Orieux Y, Duval Y, Tregear JW. Functional characterization of MADS box genes involved in the determination of oil palm flower structure. Journal of Experimental Botany. 2007 doi: 10.1093/jxb/erl263. in press. [DOI] [PubMed] [Google Scholar]
- Ainsworth C. Boys and girls come out to play: the molecular biology of dioecious plants. Annals of Botany. 2000;86:211–221. [Google Scholar]
- Ainsworth C, Crossley S, Buchanan-Wollaston V, Thangavelu M, Parker J. Male and female flowers of the dioecious plant sorrel show different patterns of MADS box gene expression. The Plant Cell. 1995;7:1583–1598. doi: 10.1105/tpc.7.10.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences of the USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwee SS, Van der Linden CG, Van der Schoot J, de Folter S, Angenent GC, Cheah S-C, et al. Characterization of oil palm MADS box genes in relation to the mantled flower abnormality. Plant Cell, Tissue and Organ Culture. 2006;85:331–344. [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Molecular Cell. 2000;5:569–579. doi: 10.1016/s1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- Angenent GC, Colombo L. Molecular control of ovule development. Trends in Plant Science. 1996;1:228–232. [Google Scholar]
- Barfod AS, Uhl NW. Floral development in Aphandra (Arecaceae) American Journal of Botany. 2001;88:185–195. [PubMed] [Google Scholar]
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Becker A, Winter KU, Meyer B, Saedler H, Theissen G. MADS-box gene diversity in seed plants 300 million years ago. Molecular Biology and Evolution. 2000;17:1425–1434. doi: 10.1093/oxfordjournals.molbev.a026243. [DOI] [PubMed] [Google Scholar]
- Beinaert A. Introduction à la biologie florale du palmier à huile (Elaeis guineensis Jacquin) Bruxelles: Institut National pour l'Etude Agronomique du Congo Belge; 1935. Série Scientifique no. 5. [Google Scholar]
- Benedito VA, Visser P, van Tuyl JM, Angenent GC, de Vries, SC, Krens FA. Ectopic expression of LLAG1, an AGAMOUS homologue from lily (Lilium longiflorum Thunb.) causes floral homeotic modifications in Arabidopsis. Journal of Experimental Botany. 2004;55:1391–1399. doi: 10.1093/jxb/erh156. [DOI] [PubMed] [Google Scholar]
- Chung Y-Y, Kim S-R, Kang H-G, Noh Y-S, Park M-C, Finkel D, et al. Characterization of two rice MADS box genes homologous to GLOBOSA. Plant Science. 1995;109:45–56. [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Corley RHV, Gray BS. Growth and morphology. In: Corley RHV, Hardon JJ, Wood BJ, editors. Vol. 1. Amsterdam: Elsevier; 1976. [Google Scholar]
- Corley RHV, Lee CH, Law LH, Wong CY. Abnormal development in oil palm clones. The Planter, Kuala Lumpur. 1986;62:233–240. [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences of the USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mason DA, Stolte W, Tisserat B. Floral development in Phoenix dactylifera. Canadian Journal of Botany. 1982;60:1439–1446. [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Dransfield J, Uhl NW. Palmae. In: Kubitzki K, editor. Families and genera of vascular plants, flowering plants: monocotyledons. Vol. 4. Berlin, Germany: Springer-Verlag; 1998. pp. 306–389. [Google Scholar]
- Dransfield J, Uhl NW, Asmussen CB, Baker WJ, Harley MM, Lewis CE. A new phylogenetic classification of the palm family, Arecaceae. Kew Bulletin. 2005;60:559–569. [Google Scholar]
- Dransfield J, Uhl NW, Baker WJ, Harley MM, Asmussen C, Lewis CE. Genera Palmarum. 2nd edn. London: Royal Botanic Gardens, Kew (in press); 2007. [Google Scholar]
- Durand-Gasselin T, Guen VL, Konan E, Duval Y. Oil palm (Elaeis guineensis Jacq.) plantations in Côte d'Ivoire obtained through in vitro culture – first results. Oléagineux. 1990;45:1–11. [Google Scholar]
- Egea Gutierrez-Cortines M, Davies B. Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends in Plant Science. 2000;5:471–476. doi: 10.1016/s1360-1385(00)01761-1. [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, et al. MADS-box protein complexes control carpel and ovule development in Arabidopsis. The Plant Cell. 2003;15:2603–2611. doi: 10.1105/tpc.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes and Development. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Govaerts R, Dransfield J. World checklist of palms. London: Royal Botanic Gardens, Kew; 2005. [Google Scholar]
- Hama E, Takumi S, Ogihara Y, Murai K. Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta. 2004;218:712–20. doi: 10.1007/s00425-003-1157-6. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Klenz JE, Haughn GW. UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta. 2006;223:769–778. doi: 10.1007/s00425-005-0138-3. [DOI] [PubMed] [Google Scholar]
- Jouannic S, Argout X, Lechauve F, Fizames C, Borgel A, Morcillo F, et al. Characterization of expressed sequence tags in oil palm (Elaeis guineensis Jacq.) FEBS Letters. 2005;579:2709–2714. doi: 10.1016/j.febslet.2005.03.093. [DOI] [PubMed] [Google Scholar]
- Kanno A, Seaki H, Kameya T, Saedler H, Theissen G. Heteropic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana) Plant Molecular Biology. 2003;52:831–841. doi: 10.1023/a:1025070827979. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA. Genetic basis for innovations in floral organ identity. Journal of Experimental Zoology (Molecular and Developmental Evolution) 2005;304B:526–535. doi: 10.1002/jez.b.21046. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Di Stilio VS, Schluter PM. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. International Journal of Plant Science. 2003;164:1–11. [Google Scholar]
- Kumar Agrawal G, Abe K, Yamazaki M, Miyao A, Hirochika H. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Molecular Biology. 2005;59:125–135. doi: 10.1007/s11103-005-2161-y. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Shimamoto K. Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant and Cell Physiology. 2002;43:130–135. doi: 10.1093/pcp/pcf010. [DOI] [PubMed] [Google Scholar]
- Levy YY, Dean C. The transition to flowering. The Plant Cell. 1998;10:1973–1989. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends in Plant Science. 2005;10:1360–1385. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. Genetic and molecular mechanisms of pattern formation in Arabidopsis flower development. Journal of Plant Research. 1998;111:233–242. [Google Scholar]
- Münster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene. 2001;262:1–13. doi: 10.1016/s0378-1119(00)00556-4. [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H-Y, Sakai H, et al. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development. 2003;130:705–718. doi: 10.1242/dev.00294. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Fukuda T, Nakano M, Hasebe M, Kameya T, Kanno A. The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. Orientalis (Agapanthaceae) flowers. Plant Molecular Biology. 2005;58:435–445. doi: 10.1007/s11103-005-5218-z. [DOI] [PubMed] [Google Scholar]
- Ochiai T, Nakamura T, Mashiko Y, Fukuda T, Yokoyama J, Kanno A, et al. The differentiation of sepal and petal morphologies in Commelinaceae. Gene. 2004;343:253–262. doi: 10.1016/j.gene.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Genetic analyses of signalling in flower development using Arabidopsis. Plant Molecular Biology. 1994;26:1357–1377. doi: 10.1007/BF00016480. [DOI] [PubMed] [Google Scholar]
- Park JH, Ishikawa Y, Yoshida R, Kanno A, Kameya T. Expression of AODEF, a B-functional MADS-box gene, in stamens and inner tepals of the dioecious species Asparagus officinalis L. Plant Molecular Biology. 2003;51:867–875. doi: 10.1023/a:1023097202885. [DOI] [PubMed] [Google Scholar]
- Park JH, Ishikawa Y, Ochiai T, Kanno A, Kameya T. Two GLOBOSA-like genes are expressed in second and third whorls of homochlamydeous flowers in Asparagus officinalis L. Plant and Cell Physiology. 2004;45:325–332. doi: 10.1093/pcp/pch040. [DOI] [PubMed] [Google Scholar]
- Parveez GK, Masri MM, Zainal A, Majid NA, Yunu AM, Fadilah HH, et al. Transgenic oil palm: production and projection. Biochemical Society Transactions. 2000;28:969–972. [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Rao V, Donough CR. Preliminary evidence of a genetic cause for the floral abnormalities in some oil palm ramets. Elaeis. 1990;2:199–207. [Google Scholar]
- Rudall PJ, Abranson K, Dransfield J, Baker W. Floral anatomy in Dypsis (Arecaceae–Areceae): a case of complex synorganization and stamen reduction. Botanical Journal of the Linnean Society. 2003;143:115–133. [Google Scholar]
- Savolainen V, Chase MW. A decade of progress in plant molecular phylogenetics. Trends in Genetics. 2003;19:717–724. doi: 10.1016/j.tig.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Skipper M. Genes from the APETALA3 and PISTILLATA lineages are expressed in developing vascular bundles of the tuberous rhizome, flowering stem and flower primordia of Eranthis hyemalis. Annals of Botany. 2002;89:83–88. doi: 10.1093/aob/mcf009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper M, Johansen LB, Pedersen KB, Frederiksen S, Johansen BB. Cloning and transcription analysis of an AGAMOUS- and SEEDSTICK ortholog in the orchid Dendrobium thyrsiflorum (Reichb. f.) Gene. 2006;366:266–274. doi: 10.1016/j.gene.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Southerton SG, Marshall H, Mouradov A, Teasdale RD. Eucalypt MADS-box genes expressed in developing flowers. Plant Physiology. 1998;118:365–372. doi: 10.1104/pp.118.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer FW, Rutishauser R, Endress PK. Morphology and development of female flowers in Geonoma interrupta (Arecaceae) American Journal of Botany. 2002;89:220–229. doi: 10.3732/ajb.89.2.220. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, et al. A short history of MADS-box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Tomlinson PB. The structural biology of palms. Oxford, UK: Oxford Scientific Publications; 1990. [Google Scholar]
- Tomlinson PB, Moore HE. Inflorescence in Nanorrhops ritchiana (Palmae) Journal of the Arnold Arboretum. 1968;49:16–34. [Google Scholar]
- Tzeng TY, Yang CH. A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant and Cell Physiology. 2001;42:1156–1168. doi: 10.1093/pcp/pce151. [DOI] [PubMed] [Google Scholar]
- Tzeng T, Cheng HY, Yang CH. Ectopic expression of carpel-specific MADS-box genes from lily and lisianthus causes similar homeotic conversion of sepal and petal in Arabidopsis. Plant Physiology. 2002;130:1827–1836. doi: 10.1104/pp.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl NW. Developmental studies in Ptychosperma (Palmae). I. The inflorescence and flower cluster. II. The staminate and pistillate flowers. American Journal of Botany. 1976;63:82–109. [Google Scholar]
- Uhl NW. Floral organogenesis in palms. In: Leins P, Tucker SC, Endress PK, editors. Aspects of floral development. Berlin, Germany: J. Cramer; 1988. pp. 25–44. [Google Scholar]
- Uhl NW, Dransfield J. Development of the inflorescence, androecium and gynoecium with reference to palms. In: White RA, Dickinson WC, editors. Contemporary problems in plant anatomy. New York: Academic Press; 1984. pp. 397–449. (NY). [Google Scholar]
- Uhl NW, Moore HE. Centrifugal stamen initiation in phytoelephantoid palms. American Journal of Botany. 1977;64:1152–1161. [Google Scholar]
- Uhl NW, Moore HE. The structure of the acervulus, the flower cluster of chamaedoreoid palms. American Journal of Botany. 1978;65:197–204. [Google Scholar]
- Uhl NW, Moore HE. Androecial development in six polyandrous genera representing five major groups of palms. American Journal of Botany. 1980;45:57–75. [Google Scholar]
- Van Heel WAV, Breure CJ, Menendez T. The early development of inflorescences and flowers of oil palm (Elaeis guineensis Jacq.) seen through the scanning electron microscope. Blumea. 1987;32:67–78. [Google Scholar]
- Van Tunen AJ, Eikeboom W, Angenent G. Floral organogenesis in Tulipa. Flower Newsletter. 1993;16:33–38. [Google Scholar]
- Weigel D. The genetics of flower development: from floral induction to ovule morphogenesis. Annual Review of Genetics. 1995;29:19–39. doi: 10.1146/annurev.ge.29.120195.000315. [DOI] [PubMed] [Google Scholar]
- Winter KU, Weiser C, Kaufmann K, Bohne A, Kirchner C, Kanno A, et al. Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Molecular Biology and Evolution. 2002;19:587–596. doi: 10.1093/oxfordjournals.molbev.a004118. [DOI] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, DePamphilis CW, Ma H, Theissen G. To B or Not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. Journal of Heredity. 2005;96:225–240. doi: 10.1093/jhered/esi033. [DOI] [PubMed] [Google Scholar]
- Zanis M, Soltis PS, Qiu Y-L, Zimmer EA, Soltis DE. Phylogenetic analyses and perianth evolution in basal angiosperms. Annals of the Missouri Botanical Garden. 2003;90:129–150. [Google Scholar]