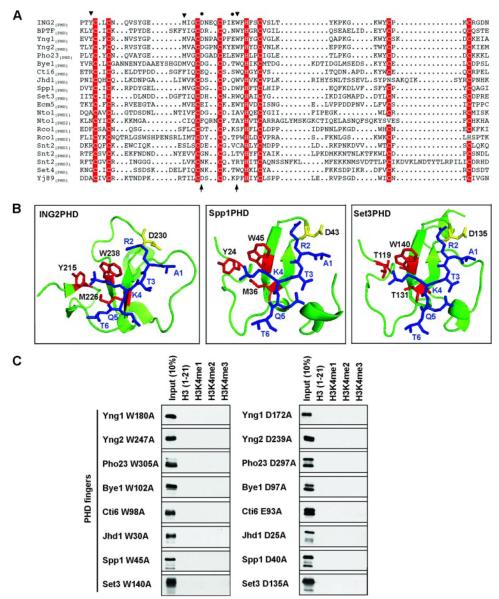
FIGURE 3. Molecular features of H3K4me3 binding by S. cerevisiae PHD fingers.
A, sequence alignment of PHD fingers from S. cerevisiae with those from human ING2 and BPTF. Zinc-coordinating residues are shaded in red; residues in the ING2 PHD finger essential for interaction of methyl-H3K4 (triangles) and H3R2 (circles) are indicated. Note that BPTF(PHD) utilizes an extra tyrosine to form a K4me-binding cage (18). Residues substituted for the mutagenesis studies are indicated with arrows at the bottom of the alignment. Sequence alignment was produced with the ESPript web server. B, structural models of Spp1(PHD) and Set3(PHD) complexed with H3(1–6)K4me3 peptide. Blue, histone peptide; red, residues that form methyl-K4 binding cage; yellow, residues essential for H3R2 interaction. Structure of the ING2(PHD)-H3K4me3 complex is shown for comparison. C, peptide pulldown assay with the indicated PHD finger mutants (left, W>A; right, D>A) performed as described for Fig. 2A.