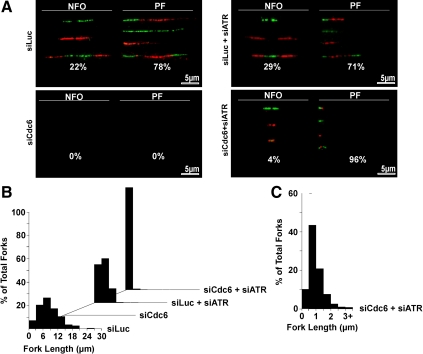
Figure 5.
Cdc6 deficiency in nontransformed cells results in ATR-mediated checkpoint suppression of progressing forks. (A) Dual-labeled (green, CldU and red, IdU) DNA fibers were generated from asynchronously growing RPE1 cells treated with indicated siRNA(s). Representative labeled DNA fibers and percentage of total DNA fibers for new firing origins (NFO) or progressing forks (PF) for each condition are shown. We counted ≥950 labeled DNA fibers from three independent experiments for each condition. (B) Lengths of all counted replication forks were measured by ImageJ (National Institutes of Health), sorted into increasing 3-μm bins, and plotted as percentage of total fork histograms on Excel (Microsoft). (C) Histogram of increased bin resolution (0.5-μm bins) showing a more detailed replication fork length distribution of siCdc6- and siATR-cotreated RPE1 cells.