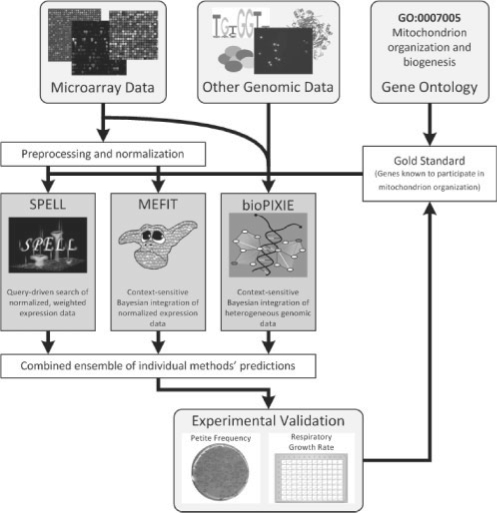
Fig. 1.
Overview of the system employed for computational function prediction and medium-throughput experimental validation. We used three computational data integration systems to predict S.cerevisiae genes functioning in the area of mitochondrion biogenesis. An initial gold standard was generated from the GO and used to train two of the machine learning systems: MEFIT, which integrates microarray data, and bioPIXIE, which integrates other diverse genomic data. SPELL was queried using mitochondrial genes from the same gold standard. Genes predicted to function in mitochondrion biogenesis after training or as the result of queries were combined and used to select candidate genes for experimental validation. Genes that significantly perturbed mitochondrion biogenesis when deleted were added to the gold standard, the three prediction methods were retrained, and a second round of experimental validation was performed. In addition to discovering many genes not previously known to participate in mitochondrial biogenesis, this process revealed striking discrepancies between the computational methods' abilities to predict experimental results and their apparent performance based on standard machine learning cross-validation.