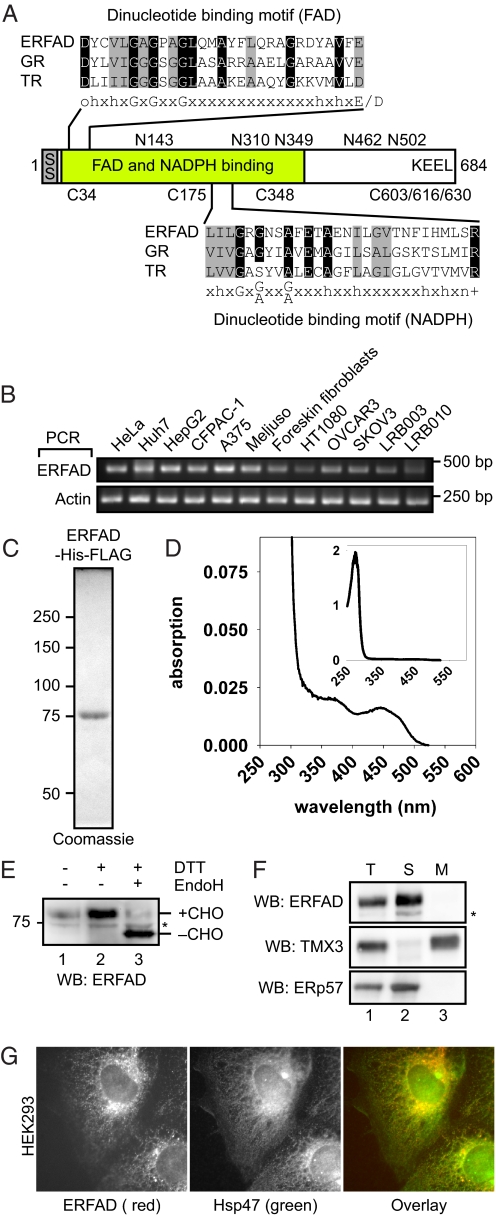
Fig. 1.
ERFAD is an ER flavoprotein. (A) Domain organization of the ERFAD protein. The two dinucleotide-binding motifs of the GXGXXG-type for FAD and NADPH binding are shown aligned with the corresponding motifs in GR, TR, and a consensus motif (amino acid residues: h = hydrophobic, o = polar/charged, + = positively charged, n = neutral). The sequence positions of the five N-glycosylation sites and the six cysteines in ERFAD are depicted. SS, signal sequence. (B) RT-PCR analysis of ERFAD. Total RNA was isolated from different human tissue culture cells, reverse transcribed and amplified with primers specific for ERFAD and actin. HeLa: cervical epithelial carcinoma; Huh7, HepG2: hepatocellular carcinoma; CF-PAC-1: pancreatic adeno carcinoma; A375, Meljuso: melanoma; HT1080: fibrosarcoma breast cancer; OVCAR3, SKOV3: ovarian epithelial carcinoma; LRB003, LRB010: embryonic stem cells (C) Purified recombinant ERFAD-His-FLAG visualized by Coomassie staining. (D) Absorption spectra of purified ERFAD-His-FLAG. The two peaks at 370 nm and 450 nm are indicative of the flavin cofactor. (Inset) Complete spectrum including the protein peak at 280 nm. (E) Glycosylation and oxidation state of human ERFAD. Lysates from HEK293 cells were treated as indicated, and analyzed by Western blotting against endogenous ERFAD. *, background band; CHO, N-glycans. (F) Subcellular fractionation of HEK293 cells. After isolation and sodium carbonate extraction of crude membranes, followed by ultracentrifugation through a sucrose cushion, the distribution of ERFAD, ERp57 (a soluble ER protein) and TMX3 (an ER membrane protein) was visualized by Western blot analysis. *, background band. (G) Immunofluorescence microscopy of ERFAD in HEK293 cells. Cells were fixed and stained with anti-ERFAD (Left, 1F6, red) and anti-Hsp47 (Center, green). A merged image is shown in the Right.