Figure 3. Gel shift of recombinant zfs1 with ARE-containing RNA oligonucleotides.
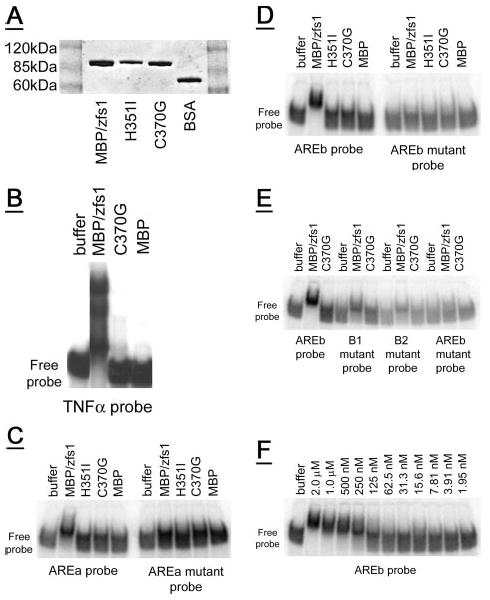
A, a Coomassie Blue-stained SDS-PAGE gel showing recombinant MBP/zfs1 (0.6 μg) compared with recombinant MBP/zfs1 zinc finger mutants (C370G, 0.5 μg, and H351I, 0.4 μg), and albumin (bovine serum albumin, 0.5 μg). Molecular weight marker positions are indicated. B, MBP/zfs1 completely shifted the tumor necrosis factor-α-based probe into the gel, whereas neither the C370G amino acid mutant of MBP/zfs1, nor MBP alone, caused the same gel shift. C, a partial shift is observed for an arz1 AREa-based probe with MBP/zfs1, but not with either zinc finger mutant MBP/zfs1 protein or MBP alone. Mutations in the binding site of the AREa probe eliminated the partial shift observed with MBP/zfs1. D, an AREb-based probe is completely shifted into the gel by MBP/zfs1, but not with either zinc finger mutant MBP/zfs1 protein or MBP alone. Mutations in the two binding sites of the AREb probe eliminated the shift observed with MBP/zfs1. E, mutations in either AREb binding site (B1 or B2) resulted in a partial shift. 50 pmol of each protein was assayed. F, a serial dilution of 50 pmol of MBP/zfs1 was used in the gel shift assay with the AREb probe, resulting in the range of concentrations shown. Half-maximal binding of the AREb probe (100 fmol total, 4 nm final concentration) was observed with ∼3.125–6.25 pmol of MBP/zfs1 (125–250 nm final concentration). For each gel (B–F) the position of the free probe at the bottom of each gel is indicated.
