Figure 4. Comparison of the zfs1 TZF domain to other TTP family members.
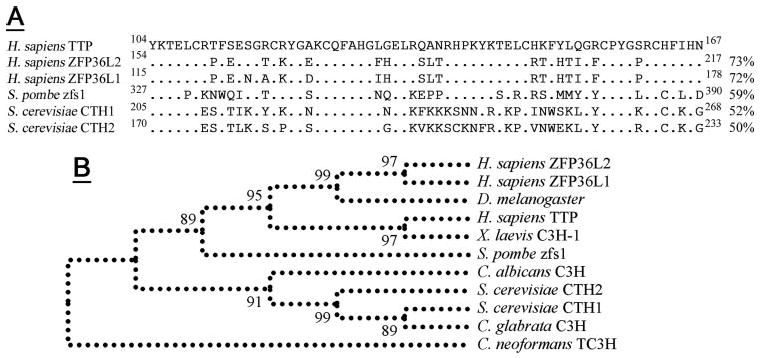
In A the zfs1 TZF is aligned with the three human TTP family members and the two family members expressed in S. cerevisiae. Percent amino acid identity with the human TTP TZF domain is shown (right) and identical residues to the human TTP sequence are indicated (dots). Conserved residues are underlined (top), with key Cys and His residues indicated (asterisks). The cladogram presented (B) shows how the zfs1 TZF domain forms a natural root to the cluster of metazoan TZFs. Other yeast TZF domains form a separate cluster, with a C. neoformans TZF domain forming a natural out-group. Bootstrap values are from 500 replicates. NCBI refseq accession numbers are as follows: Homo sapiens TTP, NP_03398.1; H. sapiens ZFP36L2, NP_008818.3; H. sapiens ZFP36L1, NP_004917.2; Xenopus laevis C3H-1, NP_001081885.1; Drosophila melanogaster Tis11, NP_511141.2; S. pombe zfs1, CAB75997.1; S. cerevisiae CTH1, P47976; S. cerevisiae CTH2, P47977; C. albicans, XP_717137.1; C. glabrata, XP_445742.1; and C. neoformans, EAL17658.1.
