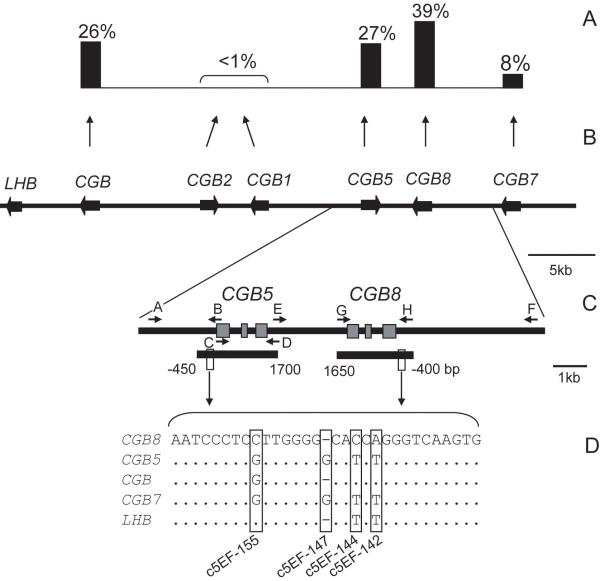
Figure 1.
Genomic and expressional context for the design of the association study targeting HCG beta genes. (A) The contribution of each individual gene into the total mRNA transcript pool of all six CGB genes (18). (B) Schematic presentation of the LHB/CGB gene cluster with genes marked as black wide arrows in the direction of transcription on sense strand. (C) The position of long-range PCR primers (black arrows) and extent of resequenced CGB5 and CGB8 regions (short black bars). Gene exons are depicted with grey boxes. Capital letters correspond to the primer sequences listed in Supplementary Table S1. (D) The aligned consensus sequences of the 5′upstream element of LHB/CGB genes. The nucleotide positions distinctive for each HCG beta and LHB gene co-localizing with CGB5 SNPs c5EF-155, c5EF-147, c5EF-144 and c5EF-142 (Table 1) are highlighted.
All positions are given relative to mRNA transcription start site.