Abstract
Despite two decades of mouse immunology and human genetics studies, the pathogenesis of Crohn's disease (CD) remains elusive. New clinical investigations suggest that CD may be caused by inborn errors of macrophages. These errors may result in impaired attraction of granulocytes to the gut wall, causing impaired clearance of intruding bacteria, thereby precipitating the formation of granulomas. This theory paves the way for a macrophage-based Mendelian genetic dissection of CD.
A heritable immunological disease
First described in 1932, CD is an inflammatory disease of the digestive tract that affects 0.1–0.2% of the adult population. Despite decades of investigation, the pathogenesis of CD has remained elusive (Baumgart and Carding, 2007). The inflammation associated with CD can occur anywhere along the route from the mouth to the anus. Although clinically defined as a digestive condition, there is no evidence that CD involves defects in epithelial or other digestive cells.
In essence, CD is an immunological disease in the hematopoietic sense of the term. This classification had long been suggested by the tissue lesions associated with CD, which consist primarily of granulomas, and by the fact that patients often respond to immunosuppressive therapy. Perhaps the best proof of an immunological etiology is the observation that CD was cured in several patients who received allogeneic hematopoietic stem cell transplantation (HSCT). Likewise, some individuals acquired CD after receiving HSCT from donors with CD. Although CD is immune mediated, it is not an autoimmune disease, as the immunological process appears to be triggered by the content of the gut lumen rather than a self-antigen.
CD is also a genetic disease, as suggested by the higher concordance in monozygotic versus dizygotic twins, the higher risk of disease in first-degree relatives of probands, and the high rate of family history among patients. However, CD does not follow a uniform mode of inheritance, a finding that has been conservatively interpreted to suggest a polygenic predisposition at both the individual and population levels (Baumgart and Carding, 2007; Van Limbergen et al., 2009). To date, CD is widely accepted as a genetically determined immunological disease that manifests principally in the gut and is triggered by bowel contents. But this is as far as scientific consensus and certainty go.
Progress and pitfalls of CD research
The immunological basis of CD has been studied primarily in various mouse models of colitis. Most of these models have suggested that colitis, and by inference CD, are mediated by T lymphocytes (Ostanin et al., 2009). In the late 1990s, human genome-wide linkage (GWL) studies were performed, followed by genome-wide association (GWA) studies in the late 2000s. Interestingly, the locus with the most robust association with CD and the first gene identified by GWL studies, NOD2/CARD15, was expressed in the myeloid rather than the lymphoid lineage (Hugot et al., 2001; Ogura et al., 2001).
NOD2 is a cytosolic receptor that recognizes bacterial peptidoglycans, particularly mycobacterial N-glycolyl muramyl dipeptide (Coulombe et al., 2009). After the identification of NOD2 as a CD-associated gene, various immunological scenarios were imagined. Some studies claimed that the CD-predisposing NOD2 mutant alleles were hypomorphic, others that they were hypermorphic. Most, however, attributed CD pathogenesis to NOD2-mediated recognition of bacterial peptidoglycan and subsequent activation of the NF-κB pathway. The observation that other alleles of NOD2 caused a clearly distinct, remotely related, and highly penetrant Mendelian autosomal dominant systemic disorder called Blau syndrome was left unexplained (Miceli-Richard et al., 2001).
Only recently did a superb study reveal the surprising fact that CD-predisposing NOD2 mutant proteins, unlike their wild-type counterparts, actively and specifically suppress the transcription of IL-10 in a peptidoglycan- and NF-κB–independent manner (Noguchi et al., 2009). This phenomenon was restricted to human NOD2 and IL-10 and was not found in their mouse orthologues, a distinction that highlights the necessity of human immunological studies (Casanova and Abel, 2004; Davis, 2008; Hayday and Peakman, 2008).
Other loci were identified by GWL studies, but the impact of these loci on disease was lower and, like NOD2, they were not universal CD susceptibility loci. Moreover, these loci were not found in GWA studies, which instead identified yet another set of loci. Altogether, up to 32 CD candidate loci have been identified by GWA studies and replicated in at least two population samples (Barrett et al., 2008). However, for none of these loci do we have a fine characterization of the immunological impact of the CD-predisposing alleles. NOD2 provides us with a preview of how long and arduous a path this is likely to be (Noguchi et al., 2009).
Missing heritability and missing intelligibility
Paradoxically, experimental difficulties in studying CD-associated alleles may not be the most challenging roadblock ahead. A more difficult issue is that the myriad of identified loci do not account for the heritability of CD, a problem referred to as missing heritability (Maher, 2008). Even given the unproven assumptions that these loci are individually causal and collectively additive, the relative risk conferred by each locus is very small, and their overall contribution would account for only 20% of CD heritability. Even for the three main CD-associated NOD2 variants, the penetrance—or the proportion of homozygotes/compound heterozygotes who develop disease—is <2%. And out of 1,000 CD patients, the number of homozygotes/compound heterozygotes and heterozygotes is relatively low, at an estimated 80 and 270, respectively, as compared with 6 and 145 in a healthy population (Hugot et al., 2007). Moreover, NOD2 was found to have no role in some populations in which CD is endemic, such as the Japanese.
An equally important problem with these genetic data is that they do not add up to a unified or plausible immunological scenario for the pathogenesis of CD. It is difficult to make immunological sense of these multiple loci and pathways, at either the cellular or the molecular level. In addition to missing heritability, and perhaps even more troubling, is the problem of missing intelligibility. Why would CD, which is epidemiologically, clinically, and pathologically definable, not obey some understandable causal relationship at the cellular, individual, and population levels? Missing intelligibility strongly suggests that the phenomenon has not been correctly understood. And if the immunological basis of CD is scientifically intelligible, one can also hope that this knowledge would be helpful to patients. This is unlikely to be the case if CD is the result of a myriad of genetic polymorphisms in various genes involved in multiple pathways, each conferring a very modest relative risk. What is more likely is that missing heritability and missing intelligibility are two sides of the same coin that account for the poor clinical impact of past investigations.
Deficit drives excess inflammation
The first breakthrough toward an understanding of the pathogenesis of CD came in 1976 when Anthony Segal demonstrated impaired influx of granulocytes into skin windows in CD patients (Segal and Loewi, 1976), a finding that was confirmed and extended to the gut in 2006 (Marks et al., 2006). As CD is an inflammatory disease, it was hard to imagine a more paradoxical and provocative finding than impaired inflammation (Korzenik, 2007). Moreover, although CD is principally localized to the gut, these studies claimed that the inflammatory defect was systemic and affected various tissues, such as the skin, that were not affected clinically. However, as the Hungarian physiologist Albert Szent-Györgyi pointed out, “a discovery is a discovery because it is at variance with accepted knowledge.”
A discovery should also provide an explanation for the phenomenon studied. In a clinical investigation reported by Smith et al. (2009) in this issue, Segal's group studied inflammation in response to skin wounded with heat-killed bacteria. Consistent with their past studies, patients with CD had impaired inflammation at the wound site, as demonstrated by tracking intravenously injected, radio-labeled granulocytes. Clearance of Escherichia coli at the sites of injection was also impaired in patients with CD, but not in those with another inflammatory disease, ulcerative colitis, or in healthy subjects. Notably, impaired bacterial clearance was evident for high, but not low, bacterial doses. Overall, these experiments suggest a causal link between impaired inflammation and impaired bacterial clearance. The intestine was not wounded with bacteria, but the skin defect was proposed to be systemic and to involve the gut in particular. In fact, E. coli was chosen for these studies because of its abundance in the intestinal flora. It is important to reiterate that the defect was seen only with high bacterial loads, given that the highest bacterial load in the human body is found in the intestinal tract, which is the primary target of CD. Impaired bacterial clearance presumably results in chronic inflammatory responses, accompanied by accumulation of granulomas consisting of macrophages and T lymphocytes. Therefore, in this model, excessive inflammation is the consequence of an underlying immunodeficiency rather than the primary cause of CD pathogenesis (Fig. 1).
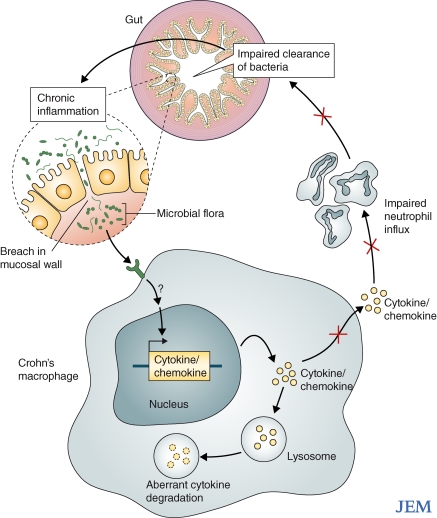
Figure 1.
A model of intestinal chronic inflammation caused by inborn errors of macrophages in patients with CD. Macrophages from the patients are intrinsically defective, with impaired secretion of cytokines that are normally translated but internally degraded. Because of insufficient production of cytokines and chemokines, there is impaired attraction of granulocytes to mucosal breaches. Impaired acute, granulocytic inflammation results in impaired clearance of bacteria and debris from the gut wall, itself resulting in chronic, granulomatous inflammation.
Tracking the culprit
That impaired inflammation hampers bacterial clearance is understandable, but what causes the impaired attraction of granulocytes to experimental wounds, and by extension, to natural mucosal breaches, in CD patients? The patients' granulocytes appear to be intrinsically intact, as they responded normally to in vitro exposure to known chemoattractants. However, macrophages derived from blood monocytes of CD patients failed to secrete proinflammatory cytokines and chemokines, including some that attract granulocytes (Smith et al., 2009). Remarkably, like the impaired attraction of granulocytes and bacterial clearance, this phenotype was shared by all CD patients tested, regardless of the NOD2 genotype in particular, and was markedly distinct from healthy controls. The defect applied to various experimental conditions, including macrophages stimulated by E. coli or microbial products.
The mechanism underlying the cytokine defect was not impaired transcription of the corresponding genes, nor was it impaired translation of the corresponding mRNAs. Instead, the cytokines appeared to be destroyed inside the cell. Treatment with lysosomal inhibitors restored cytokine production, suggesting that some normally secreted proteins were erroneously targeted to the lysosomes. In conclusion, these studies provide a plausible, unified explanation for the cellular pathogenesis of CD. This groundbreaking theory proposes that resident gut macrophages in CD patients display a phenotype marked by enhanced destruction of cytokines, which results in insufficient ignition of inflammation upon mucosal invasion by the luminal content. In turn, insufficient inflammation results in impaired clearance of bacteria and debris, leading to a T cell–mediated granulomatous response.
CD: a macrophage immunodeficiency?
The macrophage immunodeficiency theory of CD raises at least two questions. First, if the macrophage phenotype is systemic, why does CD principally occur in the gut? As suggested by the bacterial load–dependent clearance defect, the anatomical selectivity of disease may reflect the uniquely high bacterial load in the intestinal lumen, which is not found at any other interface. The bacterial content in the gastrointestinal tract is huge when compared with other locations, such as the skin or the respiratory, urinary, or genital tracts. In fact, the immunodeficiency might explain the apparently high rate of appendicitis in patients with CD (Fujita, 2009). Moreover, although the gut is the primary target of disease, immunodeficiency in patients with CD is not strictly limited to the gastrointestinal tract, as patients appear to be somewhat more prone to infectious diseases elsewhere (e.g., urinary tract infections; Kyle, 1980). A systemic macrophage defect is also consistent with the occurrence of extraintestinal manifestations of CD (Ephgrave, 2007).
If CD is a phagocyte immunodeficiency that combines a primary macrophage defect and a secondary granulocytic defect, one might expect CD to occur in patients with well-known phagocyte disorders. Indeed, CD is known to occur frequently in patients with chronic granulomatous disease (CGD), which is caused by a group of genetically determined defects of the phagocyte respiratory burst (Marks et al., 2009). Patients with CGD often present with an intestinal disease indistinguishable from CD. Other phagocyte primary immunodeficiencies (PID) also cause CD in some patients (Rahman et al., 2008). Interestingly, however, other phagocytic disorders, such as those caused by IRAK-4 and MyD88 deficiency, do not seem to predispose to CD (Ku et al., 2007; von Bernuth et al., 2008). Although patients with the latter defects are too young and too few for researchers to draw firm conclusions, this observation suggests that the CD-causing macrophage phenotype and the resulting immunodeficiency may be TLR- and IL-1R–independent.
Could CD be Mendelian?
The fact that CD patients display a macrophage immunodeficiency in turn begs the question whether the macrophage defect is truly a genetically determined, CD-causing phenotype. Ironically, we learned from GWL and GWA studies that the genetic architecture of CD may not be purely polygenic. With up to 30% familial forms and a prevalence of only ∼0.1% of the population, CD may very well result from a collection of single-gene lesions that affect macrophages and display incomplete penetrance at the clinical level, at least in a fraction of patients, including most familial and some sporadic cases. Even herpes simplex encephalitis, which is almost always sporadic, turned out to be Mendelian in at least some patients (Zhang et al., 2007).
Interestingly, even a small proportion of Mendelian forms of CD may largely explain the overall missing heritability. A simple computation shows that the contribution of Mendelian forms to the overall sibling relative risk is αRRM, where α is the proportion of Mendelian forms and RRM is the relative risk of CD in siblings of probands with Mendelian CD.
For example, if we assume an overall prevalence of CD of 1 in 1,000 (10−3) and a dominant model (heterozygotes can be affected) with a clinical penetrance of 40% for mutation carriers, the CD risk in individual siblings of probands who are heterozygous for the mutation would be 0.5 (probability of carrying the mutation) × 0.4 (clinical penetrance for mutation carriers), or 0.2. This would lead to a RRM of 200 (0.2/10−3 = 200). If we also conservatively assume that only 5% of CD cases are Mendelian (α = 0.05), a RRM of 200 leads to an estimate of the overall sibling relative risk of 10 (0.05 × 200 = 10), which is close to the observed relative risk among first-degree relatives (5–35), despite being solely the result of the Mendelian forms. As these rare Mendelian mutations could not be found by the GWA studies and may not be detected by GWL studies if there is genetic heterogeneity, this Mendelian hypothesis may, to a large extent, account for the missing heritability. Oddly enough, the genes found by GWL and GWA studies may be merely CD-modifying genes, as opposed to CD-causing genes.
CD as a macrophage primary immunodeficiency
Studies that incriminate macrophages in the pathogenesis of CD will facilitate the genetic dissection of disease. The body of work by Smith et al. (2009) has made it possible for researchers to focus on a cellular phenotype, both to select candidate genes expressed in macrophages and to test in macrophages the genes found by GW approaches in selected multiplex or consanguineous families. Regardless of the proportion of Mendelian forms of CD, and regardless of whatever genetic architecture of CD is revealed by comprehensive genetic studies, the macrophage theory and the known heritability of CD together provide a sufficient justification for referring to CD as a PID of macrophages.
This classification further expands the field of PID beyond its traditional and admittedly somewhat arbitrary phenotypic boundaries (Casanova and Abel, 2007). CD may actually turn out to be a typical PID, affecting patients whose host defense is unable to control their commensal, intestinal microbial flora. This scenario is reminiscent of the recent description of pulmonary alveolar proteinosis (PAP) in patients with granulocyte monocyte colony stimulating factor receptor mutations (Martinez-Moczygemba et al., 2008; Suzuki et al., 2008). In these patients, impaired surfactant destruction by alveolar macrophages results in a selective pulmonary phenotype, although the defect is shared by macrophages present in all tissues. There is a striking resemblance between CD and PAP, as both conditions are organ-specific manifestations of an inherited, systemic macrophage deficiency. The pivotal role of macrophages in multiple physiological processes and the diverse phenotypes related to macrophage defects found in the mouse model (Vidal et al., 2008) suggest that other human diseases, besides CD and PAP, may eventually turn out to be the result of macrophage anomalies. PIDs indiscriminately affecting various types of phagocytes have long been collected. The hunt for macrophage PIDs has just begun.
References
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. ; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease.Nat. Genet. 40:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart D.C., Carding S.R. 2007. Inflammatory bowel disease: cause and immunobiology.Lancet. 369:1627–1640 [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. 2004. The human model: a genetic dissection of immunity to infection in natural conditions.Nat. Rev. Immunol. 4:55–66 [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. 2007. Primary immunodeficiencies: a field in its infancy.Science. 317:617–619 [DOI] [PubMed] [Google Scholar]
- Coulombe F., Divangahi M., Veyrier F., de Leseleuc L., Gleason J.L., Yang Y., Kelliher M.A., Pandey A.K., Sassetti C.M., Reed M.B., Behr M.A. 2009. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide.J Exp Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M. 2008. A prescription for human immunology.Immunity. 29:835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephgrave K. 2007. Extra-intestinal manifestations of Crohn's disease.Surg. Clin. North Am. 87:673–680 [DOI] [PubMed] [Google Scholar]
- Fujita T. 2009. Is Crohn's disease associated with appendectomy or appendicitis? Am. J. Gastroenterol. 104:1324. [DOI] [PubMed] [Google Scholar]
- Hayday A.C., Peakman M. 2008. The habitual, diverse and surmountable obstacles to human immunology research.Nat. Immunol. 9:575–580 [DOI] [PubMed] [Google Scholar]
- Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O'Morain C.A., Gassull M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease.Nature. 411:599–603 [DOI] [PubMed] [Google Scholar]
- Hugot J.P., Zaccaria I., Cavanaugh J., Yang H., Vermeire S., Lappalainen M., Schreiber S., Annese V., Jewell D.P., Fowler E.V., et al. ; for the IBD International Genetics Consortium 2007. Prevalence of CARD15/NOD2 mutations in Caucasian healthy people.Am. J. Gastroenterol. 102:1259–1267 [DOI] [PubMed] [Google Scholar]
- Korzenik J.R. 2007. Is Crohn's disease due to defective immunity? Gut. 56:2–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C.L., von Bernuth H., Picard C., Zhang S.Y., Chang H.H., Yang K., Chrabieh M., Issekutz A.C., Cunningham C.K., Gallin J., et al. 2007. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity.J. Exp. Med. 204:2407–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J. 1980. Urinary complications of Crohn's disease.World J. Surg. 4:153–160 [DOI] [PubMed] [Google Scholar]
- Maher B. 2008. Personal genomes: The case of the missing heritability.Nature. 456:18–21 [DOI] [PubMed] [Google Scholar]
- Marks D.J., Harbord M.W., MacAllister R., Rahman F.Z., Young J., Al-Lazikani B., Lees W., Novelli M., Bloom S., Segal A.W. 2006. Defective acute inflammation in Crohn's disease: a clinical investigation.Lancet. 367:668–678 [DOI] [PubMed] [Google Scholar]
- Marks D.J., Miyagi K., Rahman F.Z., Novelli M., Bloom S.L., Segal A.W. 2009. Inflammatory bowel disease in CGD reproduces the clinicopathological features of Crohn's disease.Am. J. Gastroenterol. 104:117–124 [DOI] [PubMed] [Google Scholar]
- Martinez-Moczygemba M., Doan M.L., Elidemir O., Fan L.L., Cheung S.W., Lei J.T., Moore J.P., Tavana G., Lewis L.R., Zhu Y., et al. 2008. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1.J. Exp. Med. 205:2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli-Richard C., Lesage S., Rybojad M., Prieur A.M., Manouvrier-Hanu S., Häfner R., Chamaillard M., Zouali H., Thomas G., Hugot J.P. 2001. CARD15 mutations in Blau syndrome.Nat. Genet. 29:19–20 [DOI] [PubMed] [Google Scholar]
- Noguchi E., Homma Y., Kang X., Netea M.G., Ma X. 2009. A Crohn's disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1.Nat. Immunol. 10:471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease.Nature. 411:603–606 [DOI] [PubMed] [Google Scholar]
- Ostanin D.V., Bao J., Koboziev I., Gray L., Robinson-Jackson S.A., Kosloski-Davidson M., Price V.H., Grisham M.B. 2009. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade.Am. J. Physiol. Gastrointest. Liver Physiol. 296:G135–G146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman F.Z., Marks D.J., Hayee B.H., Smith A.M., Bloom S.L., Segal A.W. 2008. Phagocyte dysfunction and inflammatory bowel disease.Inflamm. Bowel Dis. 14:1443–1452 [DOI] [PubMed] [Google Scholar]
- Segal A.W., Loewi G. 1976. Neutrophil dysfunction in Crohn's disease.Lancet. 2:219–221 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Rahman F.Z., Hayee B.H., Graham S.J., Marks D.J.B., Sewell G.W., Palmer C.D., Wilde J., Foxwell B.M.J., Gloger I.S., et al. 2009. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease.J. Exp. Med. 206:1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Sakagami T., Rubin B.K., Nogee L.M., Wood R.E., Zimmerman S.L., Smolarek T., Dishop M.K., Wert S.E., Whitsett J.A., et al. 2008. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA.J. Exp. Med. 205:2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Limbergen J., Wilson D.C., Satsangi J. 2009. The genetics of Crohn's disease.Annu. Rev. Genomics Hum. Genet. 10.1146/annurev-genom-082908-150013 [DOI] [PubMed] [Google Scholar]
- Vidal S.M., Malo D., Marquis J.F., Gros P. 2008. Forward genetic dissection of immunity to infection in the mouse.Annu. Rev. Immunol. 26:81–132 [DOI] [PubMed] [Google Scholar]
- von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C.L., Chrabieh M., Mustapha I.B., Ghandil P., Camcioglu Y., et al. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency.Science. 321:691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis.Science. 317:1522–1527 [DOI] [PubMed] [Google Scholar]