Figure 4.
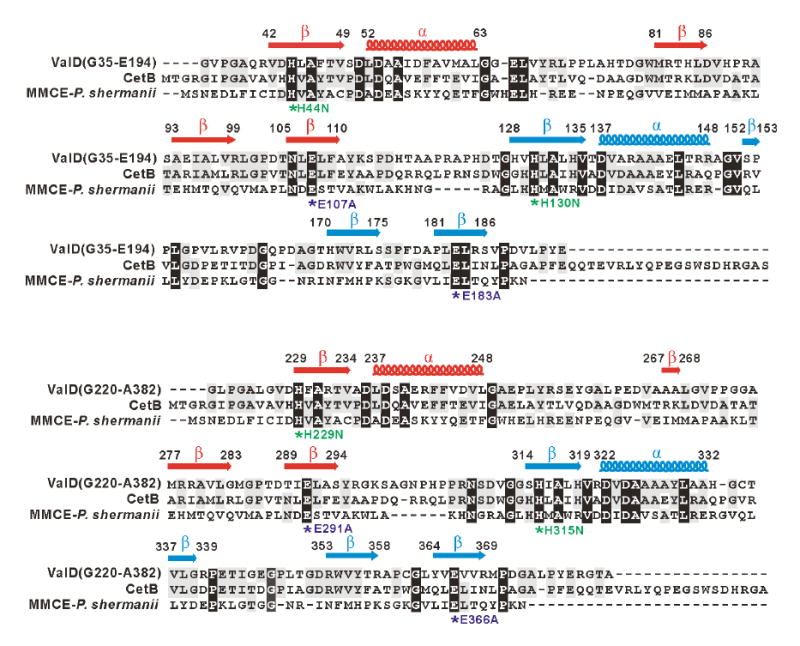
Amino acid alignments of the N-terminal half (G35-E194) and C-terminal half (G220-A382) of ValD with CetB and methylmalonyl-CoA epimerase and secondary structure predictions. Deduced amino acid sequences are from the following organisms: CetB (183 aa), Actinomyces sp. Lu 9419 (Accession No. EF120454); MMCE (148 aa), Propionibacterium shermanii (Accession No. AAL57846). Based on the analysis of the three-dimensional structure of the MMCE from Propionibacterium shermanii, a functional attribution of the aa residues probably involved in metal-ion binding and/or catalysis is indicated as asterisks below the alignment. In ValD mutants, conserved His residues are individually replaced with Asn, whereas conserved Glu residues are individually changed to Ala. The secondary structure of ValD was predicted using Jpred, and β sheets are labeled as arrows and α-helix are labeled as coils above the alignment.
