Figure 5.
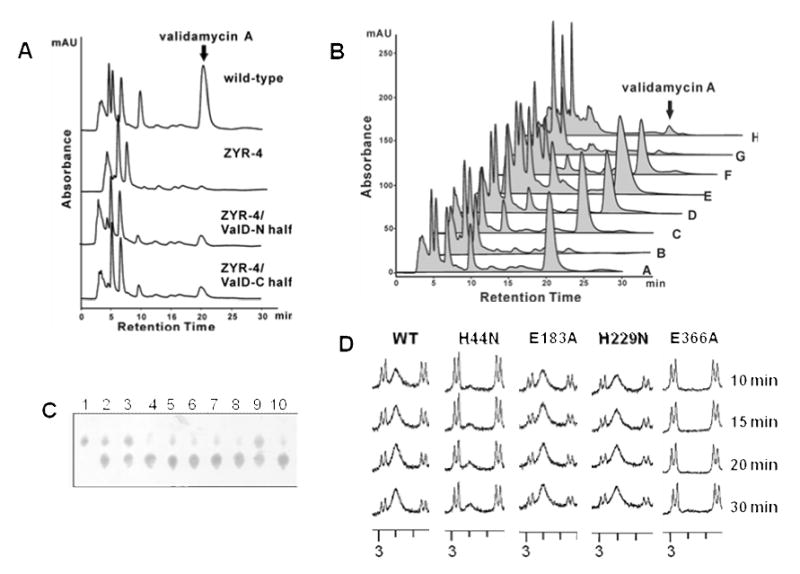
A, HPLC analysis of validamycin production in the wild-type strain, the valD disruption mutant ZYR-4, and ZYR-4 complemented with the N-terminal half and the C-terminal half of ValD. B, HPLC analysis of validamycin production in: (A) wild-type 5008; (B) valD disruption mutant ZYR-4;(C) ZYR-4 complemented with valD (H44N); (D) ZYR-4 complemented with valD (E184A); (E) ZYR-4 complemented with valD (H229N); (F) ZYR-4 complemented with valD (E366A); (G) ZYR-4 complemented with valD (H44N/H229N); (H) ZYR-4 complemented with valD (E184A/E366A). Peaks corresponding to validamycin A are indicated. C, partial TLC of enzyme assays with single-residue mutated ValD after 30 min of incubations. Lane 1, 2-epi-5-epi-valiolone standard; lanes 2 – 9, reactions with mutated proteins: H44N (2); E107A (3); H130N (4); E183A (5); H229N (6); E291A (7); H315N (8); E366A (9); 10, reaction with the wild-type ValD. D, Partial 1H NMR spectra of enzyme assays with a selected number of mutated ValD.
