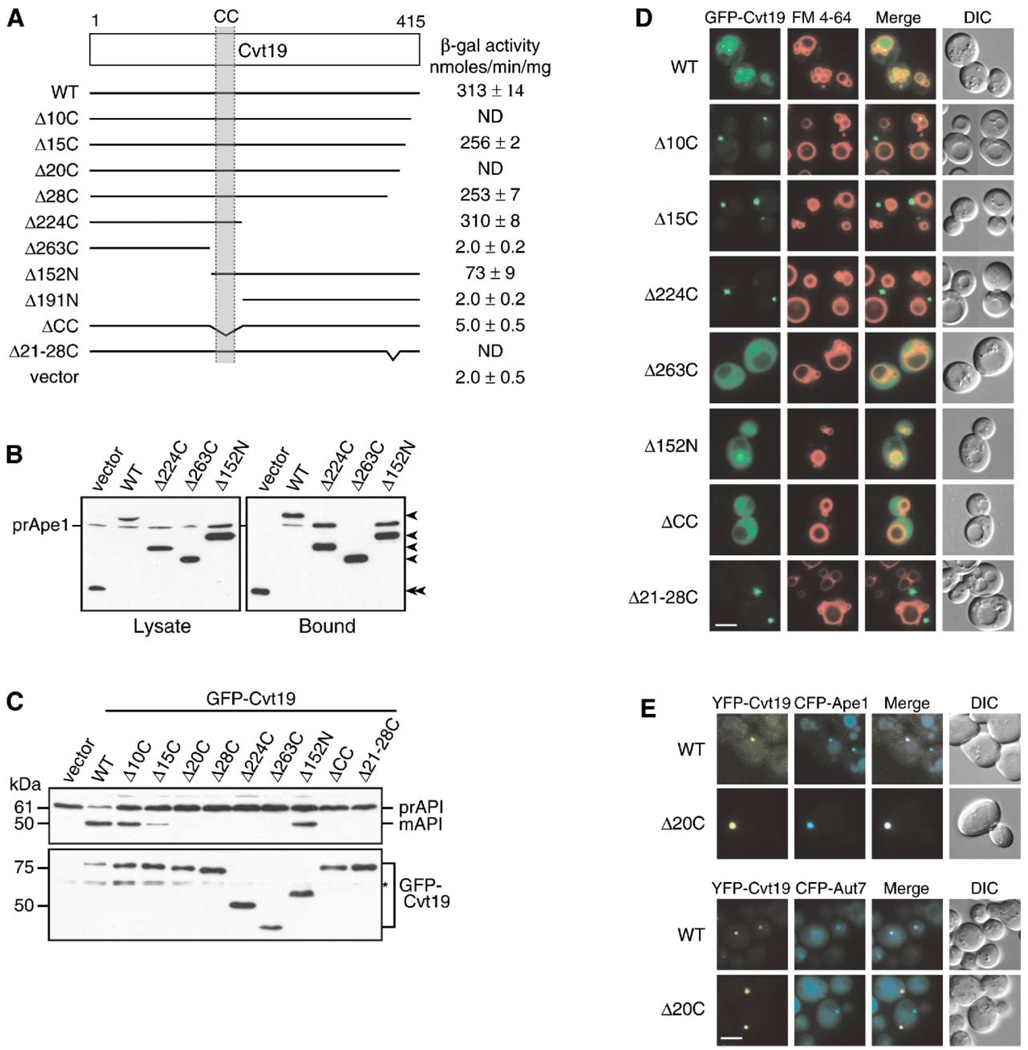
Figure 3. Mapping Functional Domains within Cvt19.
(A) Mapping the prApe1 binding domain by a yeast two-hybrid system. The cvt19Δ test strain (YTS111) was transformed with the binding domain plasmid containing the entire APE1 ORF and the activation domain plasmid containing mutant CVT19. The interaction was assessed by measuring the interaction-dependent induction of β-galactosidase activity. CC, coiled coil; ND, not determined.
(B) Total lysates from the apg1Δ cvt19Δ cells expressing either wild-type or mutant Cvt19 fused with protein A (ProtA-Cvt19) were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. The left and right panels show the protein blots of total cell extracts (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-Ape1 antiserum. Single arrowheads show ProtA-Cvt19 derivatives, and a double arrowhead indicates protein A without fusion. Although Cvt19 mutants containing the coiled-coil domain (Δ10C, Δ15C, Δ20C, Δ28C, Δ21–28C, Δ224C, and Δ152N) were all able to precipitate prApe1, only the results from Δ224C and Δ152N mutants were shown in this figure.
(C) Protein blots of total lysates from the cvt19Δ cells expressing Cvt19 deletion mutants fused with GFP probed with anti-Ape1 (upper panel) and anti-GFP (lower panel) antisera. An asterisk (*) indicates nonspecific bands.
(D) Localization of wild-type and mutant Cvt19 proteins by fluorescence and DIC microscopy of cvt19Δ strains expressing wild-type or mutant GFP-Cvt19. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm.
(E) Colocalization of YFP-Cvt19 and YFP-Cvt19Δ20C with CFP-Ape1 and CFP-Aut7 in a cvt19Δ strain by fluorescence and DIC microscopy. Bar, 5 µm.