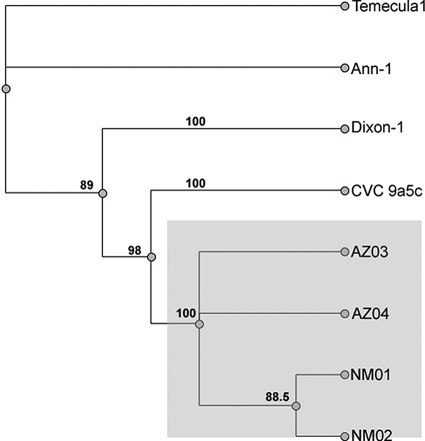
FIG. 3.
Concatameric phylogram. Sequences from eight different loci were linked for each X. fastidiosa isolate. These concatamers were used to make nucleotide alignments and phylogenetic trees using Geneious Pro v4.0.4. The maximum-likelihood method using PAUP 4.0 was utilized to make the tree. The tree was bootstrapped 1,000 times, and bootstrapping consensus percentages are shown on the tree. The concatamers included OSSR, ASSR, GSSR, gyrase-B, 272-1 (nested), RST, HL, and FY0076 sequences that were separated by four N′s to keep spatial alignment. Isolates included the New Mexico and Arizona isolates, as well as the four completely sequenced strains of X. fastidiosa CVC 9a5c (GenBank accession AE003849.1), Temecula-1 (AE009442.1), Oleander (Ann-1) (AAM03000039.1), and almond (Dixon) (AAAL02000003.1).