Abstract
Human norovirus (NoV) has been studied extensively as an important cause of gastroenteritis outbreaks worldwide. While oysters are a primary vehicle for infection, few studies have examined the wider distribution of NoV in the estuarine environment. Active shellfish-harvesting areas in Georgia were examined for the prevalence, genotype diversity, and concentrations of NoV in a variety of estuarine sample types over the course of 1 year. Of the 225 samples (9 oyster, 72 water, 72 63- to 200-μm plankton, and 72 >200-μm plankton) collected from 12 stations across two estuaries, 21 samples (9.3%) tested positive for NoV. By sample type, 55.0% (5/9) of oysters, 8.3% (6/72) of water samples, 11.1% (8/72) of 63- to 200-μm plankton samples, and 2.8% (2/72) of >200-μm plankton samples were positive for human NoV. The two NoV-positive >200-μm plankton samples, which contained mainly zooplankton, had the greatest quantity of NoV genomes (3.5 × 1013 and 1.7 × 1015 genomes g−1) of any sample tested. The majority, 90.5% (19/21), of the samples tested positive for genogroup I NoV, and only 9.5% (2/21) of the samples tested positive for genogroup II. The high concentrations of NoV in plankton samples compared to water and oyster samples were unexpected and provide new insights into the presence and distribution of human NoV in the water environment.
Human norovirus (NoV) is the leading cause of nonbacterial gastroenteritis worldwide (3). The Centers for Disease Control and Prevention (CDC) estimate that 23 million cases of acute gastroenteritis due to NoV occur each year, with symptoms including acute-onset vomiting, watery nonbloody diarrhea with abdominal cramps, and nausea (35). NoV outbreaks are pervasive for many reasons, but particularly because the virus is highly contagious and environmentally hardy (7). Additionally, infected individuals can excrete millions of viral particles in feces, leading to large numbers in sewage (16). Without proper removal or inactivation during wastewater treatment, the viruses can be released into recreational and shellfish-harvesting water bodies. Complete inactivation of NoV during sewage treatment is rare, and even in areas with proper wastewater treatment, contamination of oyster beds has been reported (5, 16, 17, 32, 38). Because bivalve molluscan shellfish are believed to act as filters for viruses and other microbes and because NoV is extremely infectious (as little as one viral particle is required for disease), the disease risk for consumption of raw oysters is high (27, 33, 40).
Human NoV genogroup I (GI) and GII have been detected in oyster samples harvested from bays and estuaries worldwide (5, 10, 20). Ueki et al. (42) detected NoV in both shellfish and the surrounding river water in Japan and concluded that NoV contamination was most likely due to sewage and treated wastewater input into the river; however, no study has yet been able to characterize how NoV may be naturally distributed in an estuarine system, including in water, adhered to particles (including plankton), and in shellfish. The limitations are due in part to a lack of adequate detection methods specifically adapted to different environmental-sample types (8). Using a newly developed detection and quantification protocol (21), this study aimed to examine the distribution of NoV genogroups across a range of sample types within an estuarine system with the goal of better characterizing possible circulation of viruses between water, plankton, and oysters.
MATERIALS AND METHODS
Controls. (i) NoV-positive controls.
Three NoV-positive fecal samples, representing genotypes GI.4, GI.3b, and GII.4 Minerva, were provided as controls for this study by the CDC. Stool samples were diluted to obtain a 20% suspension in phosphate-buffered saline (PBS), vortexed, and centrifuged at 15,700 × g for 2 min.
(ii) Viral-RNA extraction from stool.
For stool samples, viral RNA was extracted from the clarified PBS extracts using the MagMAX-96 Viral Isolation Kit (Ambion, Austin, TX) and the KingFisher Instrument (Thermo Electron Corporation, Waltham, MA), which automatically purifies viral RNA. The purified RNA was eluted into 55 μl elution buffer, provided in the kit.
(iii) RNA transcript standards.
To enable quantification by real-time reverse transcription (RT)-PCR, we used GI and GII plasmids (1) to generate RNA runoff transcripts. Briefly, the norovirus 3-kb plasmids were purified using the QIAprep Spin miniprep kit (Qiagen) and linearized with the restriction enzyme NotI (New England Biolabs, Inc., Ipswich, MA). RNA runoff transcripts were synthesized using the Megascript High Yield Transcription kit (Ambion, Austin, TX), and the transcripts were cleaned from plasmid DNA with the Megaclear kit (Ambion). Transcript integrity was confirmed by 2% agarose gel electrophoresis containing 2.2 M formaldehyde and visualized under UV light, and the transcripts were quantified spectrophotometrically at A260, diluted in diethyl pyrocarbonate-treated water to 1 × 106 copies μl−1, and stored at −80°C with 1.0 U μl−1 RNasin (Promega, Madison, WI). A 10-fold serial dilution was made (in duplicate) and used to create the standard curves for quantification.
Sample collection.
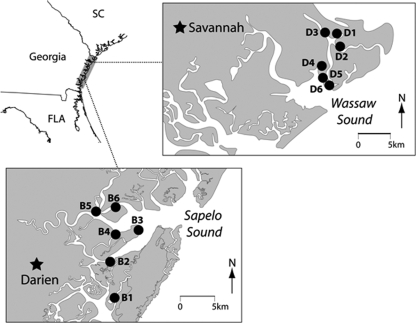
The oyster, plankton, and water samples used in this study were obtained from 12 stations representing two estuaries off the coast of Georgia. Six stations (B1 to B6) were located in Sapelo Sound, and six (D1 to D6) were located in Wassaw Sound (Fig. 1). From each station, 2 liters of water was collected and two plankton tows (for unique plankton size fractions) were performed. Oysters were not present at all stations and were, therefore, collected at only one station from each estuary, as described below. As is typical of the Georgia coast, these estuaries have large tidal ranges (1.7 to 2.1 m) but are somewhat unusual in having minimal freshwater influence from rivers (23). These estuaries had different population influences. Wassaw Sound, adjacent to Chatham County (and the city of Savannah), has a population of 241,411 (43). Sapelo Sound, near MacIntosh County, is relatively rural and has a population of 11,248 (43). Additionally, the coastal area is primarily serviced by septic systems for wastewater disposal (45), and as many as 10% of the septic systems are located within 1 ft of a water body (45). Stations B2, B3, B5, and B6 were located near a region of dense septic systems.
FIG. 1.
Sampling stations in Wassaw (stations D1 to D6) and Sapelo (stations B1 to B6) Sounds, Georgia. Water and plankton samples were collected from all stations. Oysters were collected from station D2 in Wassaw Sound and stations B3 and B5 in Sapelo Sound.
Each estuary was sampled bimonthly between September 2006 and September 2007 (no samples were collected in August 2007). Sapelo Sound was sampled in September and November 2006 and January, March, May, and July 2007. Wassaw Sound was sampled in October and December 2006 and February, April, June, and September 2007. At each sampling, the temperature, salinity, dissolved oxygen content, and pH were measured from the surface water at each station using a YSI (Yellow Springs, OH) 556 Multiparameter sonde. All samples were collected on a falling spring tide, beginning in the morning. Rainfall data for each month were obtained from the Georgia Automated Environmental Monitoring Network (http://www.georgiaweather.net) using the Skidaway Island and Brunswick stations for Wassaw and Sapelo Sounds, respectively.
(i) Oyster samples.
At each sample collection, between 20 and 40 intact oysters were collected from one public shellfish-harvesting site in each estuary. In Wassaw Sound, oysters were always collected from station D2. In Sapelo Sound, oysters were collected from station B5 from September 2006 to January 2007. High water levels prevented the collection of oysters from station B5 for the remainder of the study period. Consequently, oyster samples were collected from station B3 from February through September 2007. The oysters were kept on ice during transport and were frozen at −20°C before being processed.
(ii) Plankton samples.
Two plankton size fractions were collected using 63-μm-mesh and 200-μm-mesh plankton nets (Aquatic Research Institute, East Chicago, IN) that were towed horizontally at ∼1-m depth for 5 min. Each fraction was collected in presterilized 1-liter polypropylene bottles. The samples were held at ambient temperatures (22°C to 25°C), transported to the laboratory within 6 h of collection, and processed. Temperatures during transport were monitored with a minimum/maximum recording thermometer. Each raw plankton collection was refiltered through 63- and 200-μm nets to attain exact size fractions, 63- to 200-μm and >200-μm, and homogenized for 5 min using a Pro Scientific (Oxford, CT) Series Pro 200 homogenizer. The 63- to 200-μm fraction primarily included phytoplankton and some juvenile zooplankton, while the >200-μm fraction contained zooplankton (41). Equivalent wet weights were determined for each milliliter of sample for each fraction.
Water samples.
Two liters of water was taken just below the surface at each site in presterilized polypropylene bottles off the side of the boat. The samples were held at ambient temperature (22°C to 25°C), transported to the laboratory within 6 h of collection, and processed.
Sample processing. (i) Oysters.
NoV RNA was extracted from oyster digestive tract tissue as described previously (21). Briefly, the digestive glands of between 4 and 11 oysters, equaling at least 5 g, were removed from the shell and peripheral flesh and finely chopped with a sterile razor (22). Approximately 5 g was added to an equal volume of PBS plus 100-μg ml−1 proteinase K to degrade the shellfish tissue and release the virions into suspension. The suspension was incubated at 37°C for 1 h with shaking at 320 rpm on a C24 (New Brunswick Scientific, Edison, NJ) incubator shaker and vortexed. It was then incubated at 65°C for 15 min to inactivate the enzyme, vortexed, and centrifuged at 3,000 × g for 5 min. The soluble portion (approximately 8 ml) was aliquoted into cryovials and stored at −80°C.
One hundred and fifty microliters of supernatant from oyster homogenate was used for RNA extraction using the Qiagen RNeasy Mini Kit, following the plant and fungus procedure (14). The sample (150 μl) was added to 450 μl of a guanidine thiocyanate solution (including 45 μl β-mercaptoethanol) and vortexed vigorously. Any remaining cell debris in this suspension was removed using Qiashredder spin columns. The flowthrough was then added to 0.5 volume (∼250 μl) 100% ethanol and loaded into the RNeasy Mini columns, where bound RNA was washed with guanidine salts and ethanol and finally eluted into 50 μl nuclease-free water.
(ii) Plankton.
A plankton suspension was made, and RNA was extracted, in a method similar to that used for the oysters (21). Briefly, approximately 300 μg of homogenized plankton was separated into a microcentrifuge tube. An equal volume-to-weight of PBS plus 100 μg ml−1 proteinase K was added to the tube, and the solution was vortexed and shaken at 320 rpm for 1 hour at 37°C in a C24 (New Brunswick Scientific) incubator shaker. The tube was then heated to 65°C for 15 min to deactivate the proteinase K and centrifuged at 3,000 × g for 5 min. The supernatant (approximately 6.5 ml) was carefully removed, aliquoted, and stored at −80°C. The Qiagen RNeasy Mini kit, following the plant and fungus procedure, was used to extract RNA from 150 μl of the plankton concentrate, and the RNA was eluted into 50 μl nuclease-free water (Qiagen, Valencia, CA) (21).
(iii) Water.
The adsorption-elution method described by Katayama et al. (26) and modified by Fong et al. (15) was used to concentrate viruses from water samples. One liter of water was adjusted to a pH of ∼4.0 using a 1 N solution of acetic acid. This was passed through a 90-mm, 0.45-μm-pore-size HA membrane filter (Millipore MF Membrane Filters, Billerica, MA) using a sterile filter housing. The filter was rinsed with 100 ml 0.5 M sulfuric acid (pH 3.0). Viruses were eluted from the membrane with 10 ml 1 mM sodium hydroxide (pH 10.5 to 10.8). Eluent was added to 100 μl of 50 mM sulfuric acid (pH 3.0) and 0.1 ml of 100× Tris EDTA (pH 8.0) in a sterile 15-ml polypropylene tube. The eluent was further purified and concentrated using Centriprep YM-50 concentrator columns (Millipore, Billerica, MA) to a final volume of 2 ml. The concentrates were saved at −80°C. RNA was extracted from 200 μl of concentrate using the Qiagen RNeasy Mini kit to elute a final concentrated virus sample in 50-μl nuclease-free sterile water (Qiagen, Valencia, CA).
Detection and quantification of NoV RNA.
Two established assays for NoV real-time RT-PCR, previously evaluated for use with naturally contaminated shellfish and plankton, were utilized (Table 1) (21). The primer/probe sets described by Kageyama et al. (25) target an 84-bp fragment (GI) and a 97-bp fragment (GII) of the conserved region at the open reading frame 1-open reading frame 2 junction of the NoV genome, and the primer/probe sets described by Jothikumar et al. (24) target a 96-bp (GI) and an 89-bp (GII) fragment of the same area of the genome.
TABLE 1.
Oligonucleotide primer and probe sequences for NoV real-time RT-PCR used in this study
| Genogroup | Oligonucleotide | Sequence (5′-3′)a | Locationsb | Reference |
|---|---|---|---|---|
| GI | COGIF | CGY TGG ATG CGN TTY CAT GA | 5291-5310 | 25 |
| COGIR | CTT AGA CGC CAT CAT CAT TYA C | 5375-5358 | 25 | |
| Ring1a | FAM-AGA TYG CGA TCY CCT GTC CA-BHQ | 5340-5359 | 25 | |
| Ring1b | FAM-AGA TCG CGG TCT CCT GTC CA-BHQ | 5340-5321 | 25 | |
| GI | JJVIF | GCC ATG TTC CGI TGG ATG | 5282-5299 | 24 |
| JJVIR | TCC TTA GAC GCC ATC ATC AT | 5377-5358 | 24 | |
| JJVIP | FAM-TGT GGA CAG GAG ATC GCA ATC TC-BHQ | 5319-5341 | 24 | |
| Ring1b | FAM-AGA TCG CGG TCT CCT GTC CA-BHQ | 5340-5321 | 24 | |
| GII | COG2F | CAR GAR BCN ATG TTY AGR TGG ATG AG | 5003-5023 | 25 |
| COG2R | TCG ACG CCA TCT TCA TTC ACA | 5100-5080 | 25 | |
| Ring2 | FAM-TGG GAG GGC GAT CGC AAT CT-BHQ | 5048-5067 | 25 | |
| GII | JJV2F | CAA GAG TCA ATG TTT AGG TGG ATG AG | 5003-5028 | 24 |
| COG2R | TCG ACG CCA TCT TCA TTC ACA | 5100-5080 | 25 | |
| Ring2 | FAM-TGG GAG GGC GAT CGC AAT CT-BHQ | 5048-5067 | 25 |
The RT-PCR mixture for all assays contained 2 μl of sample, each primer at a concentration of 400 nM, each probe mixture at a concentration of 120 nM, 12.5 μl of 2× RT-PCR buffer, 1 μl of 25× RT-PCR enzyme mixture, 1.67 μl of detection enhancer, and nuclease-free water for a total reaction mixture of 25 μl (Ambion AgPath-ID One-Step RT-PCR kit). The reaction mixture was subjected to a one-step assay on an ABI 7900 (Applied Biosystems, Foster City, CA), an ABI StepOne (Applied Biosystems, Foster City, CA), or an Eppendorf (Hamburg, Germany) Mastercycler ep realplex under the following conditions: (i) RT for 10 min at 45°C, (ii) 10 min at 95°C, and (iii) 45 cycles of 10 s at 95°C, 30 s at 55°C, and 15 s at 72°C.
All amplification reactions were carried out in duplicate, resulting in a total of 8 reactions per sample (i.e., 2 primer sets × 2 genogroup targets × 2 replicates). RNA extracts from samples that showed a positive signal in either or both of the duplicate reactions were reamplified by RT-PCR to confirm the result. Only after a sample produced a second positive result was it counted as an overall positive. The mean cycle crossing-point value was calculated from the final duplicate samples and compared to standard curves created using RNA transcripts for GI.4 and GII.4 to determine the number of genome copies present (21). For simplicity, when both primer sets resulted in a positive signal for a specific genogroup target, the genome copy number was determined using a standard curve based on the primer set described by Jothikumar et al. (24). The limits of sensitivity for the two primer sets are similar and range from 20 to 2,000 genome copies, with higher detection limits noted for GII (21). Detection efficiencies following these extraction methods range from 80% for oysters to 0.14% for plankton (21); for water, this method is efficient up to 5.9% (44).
To ensure quality control, sample preparation and reagent preparation were each carried out in separated work areas, using dedicated workstations equipped with UV lamps, in a controlled-access laboratory. Samples were added to reaction tubes using only positive-displacement pipettes to avoid possible carryover contamination. A no-template negative control (nuclease-free water) was included in all runs. All amplifications took place in a physically separated laboratory with an independent air-handling system.
Statistical analyses.
Data were analyzed using SAS (Cary, NC) and Minitab (State College, PA) software for Windows. Spearman's rank correlations were performed to compare NoV concentrations with environmental parameters, including rainfall and temperature. Kruskal-Wallis tests were used to compare NoV concentrations between sample types (i.e., oysters, water, and plankton). Tukey's honest significant-difference test was used to compare the proportions of samples positive for each of the NoV genogroups (GI versus GII) across all samples (MULTPROP.MAC macro in Minitab v.14). In all cases, significance was declared at a P value of ≤0.05.
RESULTS
A total of 10 oyster samples were collected during the year; high water levels prevented collection during March and April of 2007 in Wassaw and Sapelo Sounds, respectively. Additionally, the oysters collected in October 2006 were not included in the study because they were depleted before a validated extraction method was implemented (21).
In all, 225 samples (9 oyster, 72 water, 72 63- to 200-μm plankton, and 72 >200-μm plankton) were analyzed, of which 21 samples (9.3%) were positive for NoV. The average concentrations of NoV genome copies were 2.8 × 107 (±6.6 × 107 standard deviation [SD]) g−1 for oyster samples, 1.9 × 104 (±1.6 × 105 SD) ml−1 for water samples, 1.0 × 107 (±7.6 × 107 SD) g−1 for 63- to 200-μm plankton, and 1.7.× 1015 (±2.0 × 1014 SD) for plankton >200 μm (Table 2).
TABLE 2.
Positive NoV samples among sample types
| Source | Genogroup | Estuary | Station | Date | No. of viral genomes g−1 or ml−1 | No. of genomes oyster−1a |
|---|---|---|---|---|---|---|
| Oyster | GI | Wassaw | D2 | Dec 06 | 1.1 × 104 | 1.2 × 104 |
| GI | Sapelo | B5 | Jan 07 | 8.7 × 103 | 8.7 × 103 | |
| GI | Wassaw | D2 | Feb 07 | 2.0 × 108 | 1.2 × 108 | |
| GI | Sapelo | B3 | May 07 | 9.0 × 106 | 1.1 × 107 | |
| GI | Wassaw | D2 | Jun 07 | 4.1 × 107 | 5.2 × 107 | |
| Water | GI | Wassaw | D4 | Dec 06 | 1.3 × 102 | |
| GI | Wassaw | D2 | Feb 07 | 5.2 × 102 | ||
| GI | Sapelo | B2 | Mar 07 | 2.3 × 102 | ||
| GI | Wassaw | D2 | Sep 07 | 1.4 × 106 | ||
| GII | Wassaw | D3 | Sep 07 | 1.1 × 10−1 | ||
| GI | Wassaw | D4 | Sep 07 | 3.1 × 100 | ||
| Plankton, 63-200 μm | GI | Wassaw | D3 | Feb 07 | 3.9 × 104 | |
| GI | Sapelo | B3 | Mar 07 | 9.2 × 107 | ||
| GI | Sapelo | B5 | Mar 07 | 6.4 × 108 | ||
| GI | Sapelo | B2 | May 07 | 9.5 × 103 | ||
| GII | Sapelo | B2 | May 07 | 3.9 × 104 | ||
| GI | Sapelo | B5 | May 07 | 4.4 × 104 | ||
| GI | Wassaw | D3 | Sep 07 | 3.6 × 104 | ||
| GI | Wassaw | D5 | Sep 07 | 3.7 × 106 | ||
| Plankton, >200 μm | GI | Sapelo | B3 | Mar 07 | 3.5 × 1013 | |
| GI | Sapelo | B5 | Mar 07 | 1.7 × 1015 |
Only the digestive tracts of oysters were used and weighed; thus, genomes oyster−1 can also be reported as genomes per oyster digestive tract.
Comparison by sample location.
In Wassaw Sound, of the 112 samples analyzed, 11 (10%) tested positive for NoV. Similarly, in Sapelo Sound, of 113 total samples analyzed, 10 (9%) tested positive for NoV. The average concentrations of NoV genome copies among Wassaw Sound samples were 6.0 × 107 (±9.5 × 107 SD) g−1 for oyster samples, 3.9 × 104 (±2.3 × 105 SD) ml−1 for water samples, and 1.0 × 105 (±6.2 × 105 SD) g−1 for 63- to 200-μm plankton (NoV was not detected in the >200-μm plankton samples). The average concentrations for Sapelo Sound were 1.8 × 106 genomes (±4.0 × 106 SD) g−1 for oyster samples, 6.4 genomes (±38 SD) ml−1 for water samples, 2.0 × 107 genomes (±1.1 × 108 SD) g−1 for 63- to 200-μm plankton, and 4.8 × 1013 genomes (±2.8 × 1014 SD) g−1 for >200-μm plankton.
In Wassaw Sound, 50% (5/10) of positive samples (and 25% of total positive samples, including both Wassaw and Sapelo Sounds) were detected at station D2 (oyster and water). While no station in Sapelo Sound had a statistically greater percentage of positive samples, 75% of NoV-positive samples were detected at stations B3 and B5 combined (oyster and both plankton fractions).
Temporal and seasonal distribution.
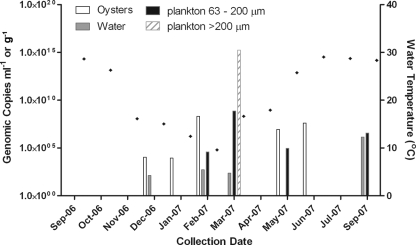
NoV-positive samples were detected in 7 of the 12 months studied (Table 2 and Fig. 2). The sample with the greatest concentration of NoV was a >200-μm plankton sample (1.7 × 1015 genomes g−1) collected in March 2007, when the highest concentration in the 63- to 200-μm fraction (6.4 × 108 genomes g−1) was also detected, both from Sapelo Sound. The largest concentration in water (1.4 × 106 genomes ml−1) occurred in September 2007, and the highest concentration in oysters (2.0 × 108 genomes g−1) occurred in February 2007, both from Wassaw Sound. When analyzed by season, 2 of 57 samples (4%) were positive for NoV in fall (October to December), 9 of 57 samples (16%) were positive in winter (January to March), 5 of 56 samples (9%) were positive in spring (April to June), and 5 of 56 samples (9%) were positive in summer (July to September).
FIG. 2.
Total NoV load (GI and GII combined) by month showing the sum of contributions from each sample type and water temperature (⧫). No samples were collected in August 2007.
The potential relationship of NoV levels to environmental parameters was also examined, and while there was an inverse association between both NoV concentration and rainfall and NoV concentration and temperature, there were no statistically significant relationships (data not shown). Additionally, there was no significant relationship with any of the other measured water quality parameters (pH, salinity, and dissolved oxygen content).
Comparison by sample type.
NoV was detected in every sample type analyzed. By sample type, 55.0% (5/9) of all oyster samples collected, 8.3% (6/72) of all water samples, 11.1% (8/72) of all 63- to 200-μm plankton samples, and 2.8% (2/72) of all >200-μm plankton samples were positive for human NoV (the two genogroups combined). Among all NoV-positive samples, 23.8% (5/21) were oyster samples, 28.6% (6/21) were water samples, 31.0% (8/21) were 63- to 200-μm plankton samples, and 9.5% (2/21) were >200-μm plankton samples.
Average NoV concentrations for GI ranged from 2.0 × 104 (±1.7 × 105 SD) copies ml−1 in water samples to 2.4 × 1013 (±2.0 × 1014 SD) copies g−1 in >200-μm plankton (Fig. 3). GII was never found in oysters or >200-μm plankton samples but averaged 1.5 × 10−3 (±1.3 × 10−2 SD) copies ml−1 in water and 5.4 × 102 (±4.6 × 103 SD) copies ml−1 in 63- to 200-μm plankton (Fig. 3).
FIG. 3.
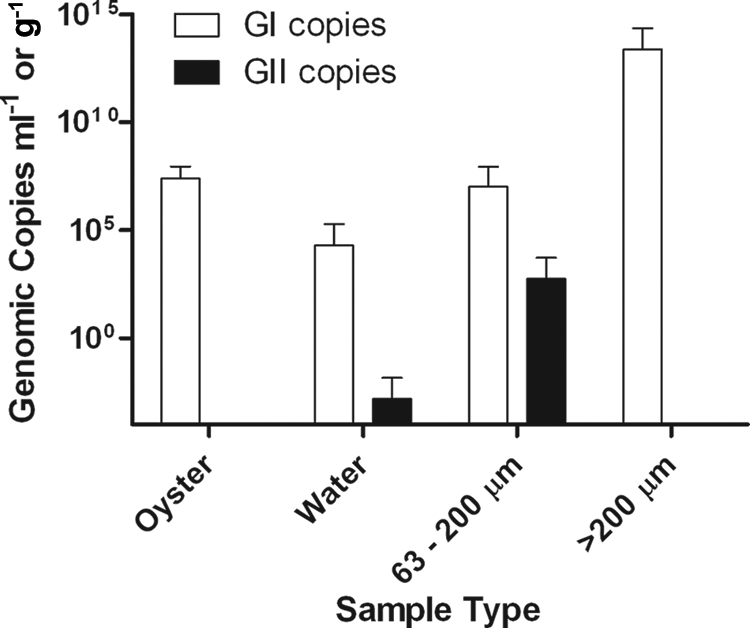
Mean NoV concentration (plus SD) for both genogroups for each sample type (oysters [n = 9], water [n = 72], 63- to 200-μm plankton fraction [n = 72], and >200-μm plankton fraction [n = 72]).
When only NoV-positive samples were considered, the concentrations for both genogroups combined ranged from a low of 1.1 × 10−1 genomes ml−1 (water; September 2007) to a high of 1.7 × 1015 genomes g−1 (>200-μm plankton; March 2007) (Table 2). Positive water samples averaged 2.3 × 105 (±5.7 × 105 SD) genomes ml−1 (n = 6). For oysters, the mean concentration among positive samples was 5.0 × 107 (±8.6 × 107 SD) genomes g−1 (n = 5), assuming all viruses were contained in the digestive tract. Among the 63- to 200-μm plankton samples, the mean concentration was 9.2 × 107 (±2.2 × 108 SD) genomes g−1 (n = 8), and the mean concentration for the >200-μm plankton samples was 8.7 × 1014 (±1.2 × 1015 SD) genomes g−1 (n = 2).
Positive findings in concurrently collected sample types (≥2) from the same station occurred four times during the study period (Table 2). NoV GI was found in oysters and water at station D2 in February 2007 and in both plankton fractions at stations B3 and B5 in March 2007. In September 2007, NoV GII was detected in water and GI was detected in the 63- to 200-μm plankton fraction at station D3.
Comparison by genogroups.
GI and GII represented 90.5% (19/21) and 9.5% (2/21) of the positive samples identified, respectively. The proportion of GI-positive samples was significantly greater than the proportion of GII-positive samples (P < 0.0001). GII NoV was detected only in a 63- to 200-μm plankton sample from Sapelo Sound in May 2007 and in a water sample from Wassaw Sound in September 2007 (Table 2).
DISCUSSION
Human NoV is the most significant viral pathogen associated with food- and waterborne outbreaks of acute gastroenteritis (3). Outbreaks due to contaminated water and oysters pose a serious risk in the United States and abroad. We investigated the distribution of NoV in shellfish-harvesting waters, a key component in the circulation of human NoV between contaminated water, food, and humans. While oysters were more often contaminated with NoV in these Georgia estuaries (55% positive) than other sample types, plankton were found to be a novel source of high virus concentrations, with up to 1.7 × 1015 genomes g−1. Additionally, NoV GI was found more frequently and at higher concentrations than GII, which is most commonly implicated in outbreaks. These results indicate that NoV, especially GI, may circulate widely between water, plankton, and oysters and suggest that accumulation by oysters could be affected by the existence of a plankton-based reservoir for these viruses in polluted waters.
Spatial trends.
Both estuaries evaluated in this study support active commercial and recreational shellfish-harvesting activities, and both were open for harvesting through the study period. The frequencies of NoV-positive samples and the average concentrations were similar in Wassaw and Sapelo Sounds, with the notable exception of very high levels detected in the >200-μm plankton fraction from Sapelo Sound. Additionally, in Sapelo Sound, 75% of positive samples were found at only two stations, both of which were situated in an area reported to contain a high density of aging septic systems (45) with inadequate drainage (23, 45). Septic systems could be a major contributor to NoV contamination in this region.
In Wassaw Sound, 5 of the 11 positive samples were recorded in September 2007 (including water and plankton at stations D2 to D5). While septic systems are also common in this region, the adjacent city of Savannah is serviced by centralized wastewater treatment. In August, the Chatham County Wastewater authority reported a minor sewage spill near Betz Creek (in the immediate vicinity of stations D2 and D3). While follow-up tests for fecal indicator bacteria suggested only a low level of contamination (data not shown), it is possible that NoV contamination in this area may reflect the influence of sewage discharge.
Seasonal trends.
Although more samples tested positive for NoV in the winter, the number was not much greater than in other seasons. This may reflect an important distinction between clinical outcome and environmental prevalence, as clinical studies in the United States and Europe report NoV primarily as a winter disease (36). Whereas NoV displays a seasonal distribution in restricted environments, such as hospitals, nursing homes, schools, and the military, the seasonal variation is much less pronounced in the larger community (34). Although studies of waterborne NoV seasonality are limited, the results of this study and others (46) suggest that NoV may be more persistent in the estuarine system over a variety of seasons than previously thought, and thus, the estuarine distribution of the virus may be an important factor in the occurrence of disease cases and outbreaks.
Distribution among estuarine sample types.
Oysters had the highest percentage of NoV-positive samples (55%) of any sample type. This is consistent with previous work showing NoV prevalence in oysters as high as 44% (12). Our detection rate may have been slightly greater due either to higher loads or to improved detection efficiency compared to earlier studies (21). Detection rates of 8% in water from this study is also consistent with the results of La Rosa et al. (30), who showed lower overall detection in natural water samples than in oysters.
NoV can persist in water and oyster samples for extended periods (37); however, the full distribution of NoV in the environment is as yet unknown. While laboratory studies show that enteric viruses are protected from microbial degradation, heat, and salts when associated with marine sediments (29), much less is known about viral association with organic particles. A novel component of this study was the examination of plankton for NoV. We found that 63- to 200-μm plankton (a mix of phytoplankton and zooplankton) (41) made up 31.0% (8/21) of all NoV-positive samples. Additionally, the >200-μm plankton (primarily zooplankton) (41) had the highest concentrations of NoV genomes of any sample. We speculate that NoV may be adsorbed to plankton particles in general via electrostatic interactions, similar to viral adsorption to sediment and other particles (28). Most enteric viruses are negatively charged under neutral and alkaline pH conditions (2). Although no work has yet characterized the charge on zooplankton particles, Bayne and Lawrence (4) found that among phytoplankton in the 6- to 100-μm size fraction, the majority of Chlorophyta, Cyanophyta, and Euglenophyta were neutral to positively charged and therefore could support adsorption of negatively charged enteric viruses. While there is no definitive evidence for the mechanism promoting virus association with plankton, it may be a significant area for future research.
If plankton, or other organic particulates, offer a reservoir for NoV, they may provide an important link for promoting virus accumulation in shellfish (hepatopancreas) tissues. Bivalves use three selective mechanisms to distinguish particles while filter feeding: particle retention, preingestive selection, and differential absorption (11). These mechanisms allow bivalves to retain only organic particles of the correct size and seston load (6). Adsorption to plankton particles could allow viral particles to be retained in the oyster and ingested in the digestive tract, whereas adsorption to sediment particles or presence in water alone could more often result in expulsion as pseudofeces. Thus, adsorption to plankton rather than inorganic (sediment) particles may be an important factor in transmission to bivalve molluscan tissues and, in turn, to humans.
Genogroup distribution.
An unexpected result of this study was that the majority of positive samples were from GI; only 9.5% of the positive samples were from GII (2 of 21 samples). The GII-positive samples also contained the smallest quantity of NoV detected in any sample throughout the study period. While both GI and GII NoVs are found commonly in sewage samples, outbreaks are more frequently attributed to GII (19, 30, 31, 39, 42). The prevalence of NoV GI strains in this study may be attributed to the use of two improved real-time assays (21), possibly giving a more accurate view of NoV GI presence in the environment; however, the limits of detection are lower for GI than for GII using these RT-PCR assays (21). Despite methodological differences, recent evidence suggests that NoV GI may be more pervasive than previously thought. For example, GI strains detected in NoV outbreaks increased from 4% between 1996 and 1997 to 25% between 1997 and 2000 according to one U.S. study (18). Additionally, among 54 U.S. travelers to Mexico and Guatemala diagnosed with traveler's diarrhea, 26 were confirmed to have NoV infections, all of which (26/26) were GI (9). An analysis of wastewater in France showed that NoV GI and GII were present in 43 and 88% of sewage influent samples, respectively, but in 24 and 14% of effluent samples, respectively, suggesting that sewage treatment is less effective for treating NoV GI than GII (13). Additionally, NoV GI was more variable, had higher peaks, and had higher average positive influent concentrations than NoV GII (13). These data suggest that there may be differences in environmental persistence between viruses belonging to the two genogroups. NoV detected in sewage and in the environment may more accurately reflect the true circulation in the population, rather than reported cases, which is a small proportion of the total cases (13).
Conclusions.
Previous studies have investigated the presence of NoV in environmental oyster samples, but few studies have examined the presence of NoV in oysters over a full year, and even fewer have examined NoV presence in water environments surrounding shellfish-harvesting areas. This is the first study to examine the association between NoV and oysters, water, and plankton. We found a higher prevalence of NoV GI than GII, suggesting that NoV GI may be more environmentally hardy than GII in the estuarine environment or that GI strains are circulating more widely in the population than expected. Additionally, we discovered that NoV is associated differentially with plankton size fractions and that the >200-μm fraction can harbor concentrations greater than 1013 copies g−1. Future research should address the notion that organic particles (i.e., plankton) can provide a reservoir for increased persistence of NoV in coastal waters. Enteric viruses concentrated within plankton fractions could feasibly increase the probability of uptake by oysters during feeding. All of these dynamics are significant new findings that point to the need for more studies of NoV in the water environment.
Acknowledgments
This work was supported by National Oceanic and Atmospheric Administration Oceans and Human Health Initiative grant no. NA04OAR4600203.
Members of the EHS Environmental Microbiology Laboratory at the University of Georgia, the members of the National Calicivirus Laboratory at the CDC, and the Georgia DNR, Coastal Resources Division (William Hughes and Brooks Good), provided valuable laboratory and field support.
None of the authors has any potential or real conflict of interest.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the funding agency or the CDC. This article received clearance through the appropriate channels at the CDC prior to submission.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. S. Noel, and R. I Glass. 1997. A one-tube method of reverse transcription-PCR to efficiently amplify a 3-kilobase region from the RNA polymerase gene to the poly(A) tail of small round-structured viruses (Norwalk-like viruses). J. Clin. Microbiol. 35:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano, T. (ed.). 1998. Wastewater reclamation and reuse, vol. 10. CRC Press, Boca Raton, FL.
- 3.Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. N. Am. 35:275-290. [DOI] [PubMed] [Google Scholar]
- 4.Bayne, D. R., and J. M. Lawrence. 1972. Separating constituents of natural phytoplankton populations by continuous particle electrophoresis. Limnol. Oceanogr. 17:481-489. [Google Scholar]
- 5.Beuret, C., A. Baumgartner, and J. Schluep. 2003. Virus-contaminated oysters: a 3-month monitoring of oysters imported to Switzerland. Appl. Environ. Microbiol. 69:2292-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bougrier, S., A. J. S. Hawkins, and M. Heral. 1997. Preingestive selection of different microalgal mixtures in Crassostrea gigas and Mytilus edulis, analysed by flow cytometry. Aquaculture 150:123-134. [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2006. Norovirus in healthcare facilities fact sheet. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/ncidod/dhqp/id_norovirusFS.html.
- 8.Centers for Disease Control and Prevention. 2006. Norovirus: technical fact sheet. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/ncidod/dvrd/revb/gastro/norovirus-factsheet.htm.
- 9.Chapin, A. R., C. M. Carpenter, W. C. Dudley, L. C. Gibson, R. Pratdesaba, O. Torres, D. Sanchez, J. Belkind-Gerson, I. Nyquist, A. Karnell, B. Gustafsson, J. L. Halpern, A. L. Bourgeois, and K. J. Schwab. 2005. Prevalence of norovirus among visitors from the United States to Mexico and Guatemala who experience traveler's diarrhea. J. Clin. Microbiol. 43:1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, P. K. C., D. K. K. Wong, T. W. H. Chung, and W. W. L. Lim. 2005. Norovirus contamination found in oysters worldwide. J. Med. Virol. 76:593-597. [DOI] [PubMed] [Google Scholar]
- 11.Cognie, B., L. Barille, and Y. Rince. 2001. Selective feeding of the oyster Crassostrea gigas fed on natural microphytobenthos assemblage. Estuaries Coasts 24:126-134. [Google Scholar]
- 12.Constanini, V., F. Loisy, L. Joens, F. S. Le Guyader, and L. J. Saif. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl. Environ. Microbiol. 72:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva, A. K., J.-C. L. Saux, S. Parnaudeau, M. Pommepuy, M. Elimelech, and F. S. L. Guyader. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups. Appl. Environ. Microbiol. 73:7891-7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Roda Husman, A. M., F. Lodder-Verschoor, H. H. J. L. Van den Berg, F. S. L. Guyader, H. V. Pelt, W. H. M. V. de Poel, and S. A. Rutjes. 2007. Rapid virus detection procedure for molecular tracing of shellfish associated with disease outbreaks. J. Food Prot. 70:967-974. [DOI] [PubMed] [Google Scholar]
- 15.Fong, T. T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formiga-Cruz, M., A. K. Allard, A. C. Conden-Hansson, K. Henshilwood, B. E. Hernroth, J. Jofre, D. N. Lees, F. Lucena, M. Papapetropoulou, R. E. Rangdale, A. Tsibouxi, A. Vantarakis, and R. Girones. 2003. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl. Environ. Microbiol. 69:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formiga-Cruz, M., G. Tofino-Quesada, S. Bofill-Mas, D. N. Lees, K. Henshilwood, A. L. Allard, A. C. Conden-Hansson, B. E. Hernroth, A. Vantarakis, A. Tsibouxi, M. Papapetropoulou, M. D. Furones, and R. Girones. 2002. Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden and the United Kingdom. Appl. Environ. Microbiol. 68:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Gallimore, C. I., M. A. Barreiros, D. W. Brown, J. P. Nascimento, and J. P. Leite. 2004. Noroviruses associated with acute gastroenteritis in a children's day care facility in Rio de Janeiro, Brazil. Braz. J. Med. Biol. Res. 37:321-326. [DOI] [PubMed] [Google Scholar]
- 20.Gallimore, C. I., J. S. Cheesbrough, K. Lamden, C. Bingham, and J. J. Gray. 2005. Multiple norovirus genotypes characterised from an oyster-associated outbreak of gastroenteritis. Int. J. Food Microbiol. 103:323-330. [DOI] [PubMed] [Google Scholar]
- 21.Gentry, J. B., J. Vinje, and E. K. Lipp. 2009. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J. Virol. Methods 156:59-65. [DOI] [PubMed] [Google Scholar]
- 22.Henshilwood, K., W. J. Dore, S. Anderson, and D. N. Lees. 2003. Investigation of Norwalk like virus elimination during depuration using a real time quantitative PCR, p. 457-465. In J. L. R. A. Villalba, B. Reura, and R. Beiras (ed.), Molluscan shellfish safety. Conselleria de Pesca e Asuntos Maritimos da Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Galicia, Spain.
- 23.Johnston, A. S., H. O. Hillestad, S. F. Shanholtzer, and G. F. Shanholtzer. 1970. An ecological survey of the coastal region of Georgia. National Park Service, Washington, DC.
- 24.Jothikumar, N., J. A. Lowther, K. Henshilwood, D. N. Lees, V. R. Hill, and J. Vinje. 2005. Rapid and sensitive detection of norovirus by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama, K., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohn, M. A., T. A. Farley, T. Ando, M. Curtis, S. A. Wilson, Q. Jin, S. S. Monroe, R. C. Baron, L. M. McFarland, and R. I. Glass. 1995. An outbreak of Norwalk virus gastroenteritis associated with eating raw oysters. Implications for maintaining safe oyster beds. JAMA 273:466-471. [DOI] [PubMed] [Google Scholar]
- 28.Labelle, R., and C. P. Gerba. 1980. Influence of estuarine sediment on virus survival under field conditions. Appl. Environ. Microbiol. 39:749-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labelle, R., and C. P. Gerba. 1982. Investigations into the protective effect of estuarine sediment on virus survival. Water Res. 16:469-478. [Google Scholar]
- 30.La Rosa, G., S. Fontana, A. Di Grazia, M. Iaconelli, M. Pourshaban, and M. Muscillo. 2007. Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 73:4152-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Guyader, F., F. Bon, D. DeMedici, S. Parnaudeau, A. Bertone, S. Crudeli, A. Doyle, M. Zidane, E. Suffredini, E. Kohli, F. Maddalo, M. Monini, A. Gallay, M. Pommepuy, P. Pothier, and F. M. Ruggeri. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J. Clin. Microbiol. 44:3878-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Guyader, F., L. Haugarreau, L. Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Guyader, F., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvoen-Clouet, M. Pommepuy, and J. Le Pendu. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopman, B. A., G. K. Adak, M. Reacher, and D. W. Brown. 2003. Two epidemiologic patterns of norovirus outbreaks: surveillance in England and Wales, 1992-2000. Emerg. Infect. Dis. 9:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181:S284-S287. [DOI] [PubMed] [Google Scholar]
- 37.Schwab, K. J., F. H. Neill, M. K. Estes, T. G. Metcalf, and R. L. Atmar. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J. Food Prot. 61:1674-1680. [DOI] [PubMed] [Google Scholar]
- 38.Shieh, Y. C., R. C. Baric, J. W. Woods, and K. R. Calci. 2003. Molecular surveillance of enterovirus and Norwalk-like virus in oysters relocated to a municipal-sewage-impacted gulf estuary. Appl. Environ. Microbiol. 69:7130-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subekti, D. S., P. Tjaniadi, M. Lesmana, C. Simanjuntak, S. Komalarini, H. Digdowirogo, B. Setiawan, A. L. Corwin, J. R. Campbell, K. R. Porter, and B. A. Oyofo. 2002. Characterization of Norwalk-like virus associated with gastroenteritis in Indonesia. J. Med. Virol. 67:253-258. [DOI] [PubMed] [Google Scholar]
- 40.Teunis, P. F. M., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. L. Pendu, and R. L. Calderon. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468-1476. [DOI] [PubMed] [Google Scholar]
- 41.Turner, J. W., B. Good, D. Cole, and E. K. Lipp. 7 May 2009. Environmental factors affect the status of plankton as a reservoir for Vibrio species. ISME J. [Epub ahead of print.] doi: 10.1038/ismej.2009.50. [DOI] [PubMed]
- 42.Ueki, Y., D. Sano, T. Watanabe, K. Akiyama, and T. Omura. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271-4280. [DOI] [PubMed] [Google Scholar]
- 43.U.S. Census Bureau. U.S. census 2000. U.S. Census Bureau, Washington, DC. http://quickfacts.census.gov/qfd/states.html.
- 44.Victoria, M., F. Guimarães, T. Fumian, F. Ferreira, C. Viera, J. P. Leite, and M. Miagostovich. 2009. Evaluation of an adsorption-elution method for detection of astrovirus and norovirus in environmental waters. J. Virol. Methods 156:73-76. [DOI] [PubMed] [Google Scholar]
- 45.Walker, R. L., C. F. Cotton, and K. A. Payne. 2003. A GIS inventory of on-site septic systems adjacent to the coastal waters of McIntosh County, Georgia. Univ. Georgia Mar. Ext. Bull. 27:1-44. [Google Scholar]
- 46.Westrell, T., P. Teunis, H. van den Berg, W. Lodder, H. Ketelaars, T. A. Stenstrom, and A. M. de Roda Husman. 2006. Short- and long-term variations of norovirus concentrations in the Meuse River during a 2-year study period. Water Res. 40:2613-2620. [DOI] [PubMed] [Google Scholar]