Abstract
Bluetongue virus (BTV) is a major pathogen of ruminants. Especially serotypes 1, 6, and 8 are of concern to veterinary authorities in central Europe. This article describes highly sensitive real-time reverse transcription-PCR assays directed to BTV genome segment 2 for specific detection of BTV-1, -6, or -8 in animal samples.
Bluetongue virus (BTV), an arthropod-borne Orbivirus in the family Reoviridae, is a major pathogen of ruminants, and there are 24 established serotypes (18). Until the end of the 20th century, bluetongue disease (BT) had been considered exotic in Europe. Since 1998, strains of at least five serotypes (BTV-1, -2, -4, -9, and -16) have been present along the Mediterranean coast (5, 14). In 2006, BTV-8 was first detected in The Netherlands without any previous notion of its presence in Europe (3). Clinical disease in sheep was severe and, unusually for BTV, it was also seen in a wide range of nonovine species, including cattle, exacerbating the economic impact (5, 13). By the end of 2008, BTV-8 was present in all of central Europe and beyond, and vaccination campaigns are ongoing (4, 6, 17). At the same time, BTV-1 of Algerian origin has expanded northward across the Iberian Peninsula and France (5), and in October 2008, BTV-6 was found in cattle in The Netherlands and later also in Germany (8, 9). Finally, in 2009, BTV-11 was reported in Flanders (10). European Union monitoring requirements and the need for fast, reliable, and sensitive detection of BTV RNA in animal samples led to the development of several real-time quantitative reverse transcription-PCR (RT-qPCR) assays (7). This article describes new assays that specifically detect BTV-1, -6, or -8 to be used for further characterization of a BTV-positive sample.
For maximum specificity, our assays are directed to BTV genome segment 2, which encodes the highly variable outer shell protein VP2 (18). VP2 sequences are available for at least one isolate of every established serotype (11; NCBI GenBank). Several independent VP2 sequences have been published for BTV-1 and -8 (12). Among these, we gave priority to sequences of European isolates. The sequence of segment 2 of NET2006/04, the European reference isolate of BTV-8, was found to have higher similarity to a 1982 isolate from Nigeria than to the South African BTV-8 reference strain (12). On the other hand, the segment 2 sequences of North African and French isolates of BTV-1 are close to that of reference strain RSArrrr/01, all belonging to the “western group” (16). For BTV-6, VP2 of the recently emerged strain is more than 99% identical to that of South African BTV-6 reference strain RSArrrr/06 (15), the only published sequence. Accordingly, primers and hydrolysis probes (Table 1) were selected by using Beacon Designer (PremierBiosoft International) and Primer3 design software (http://frodo.wi.mit.edu/). The probes were labeled with 6-carboxyfluorescein (FAM) at the 5′ end, and black hole quencher 1 carboxylic acid was attached to the 3′ end. Oligonucleotides were synthesized by a commercial supplier (Eurogentec Deutschland GmbH).
TABLE 1.
Primers and probes used in this study
| Oligonucleotide (product size [bp]) and type | Name | Sequence |
|---|---|---|
| BTV1-VP2-Mix1-FAM (123) | ||
| Forward primer | BTV1-VP2-186F | 5′ CGG ACC GCA TTA TGG TAT AAC C 3′ |
| Reverse primer | BTV1-VP2-308R | 5′ ACT CTT GTG TCT CGT ACT TTC AAC 3′ |
| Probe | BTV1-VP2-203FAM | 5′ ACC GCC CGT CTT TCA TCG TAA CCC 3′ |
| BTV1-VP2-Mix2-FAM (120) | ||
| Forward primer | BTV1-VP2-2407F | 5′ CCT CAA AGG CGA TTC GAT TTA GC 3′ |
| Reverse primer | BTV1-VP2-2526R | 5′ TCA CGA CGT TGT AGT TGA CTC C 3′ |
| Probe | BTV1-VP2-2438FAM | 5′ TGA AGC GCA GCC CAA GAT TGC ACG 3′ |
| BTV6-VP2-Mix1-FAM (97) | ||
| Forward primer | BTV6-VP2-785F | 5′ GAT ACG TGA TGC GTG GAT TG 3′ |
| Reverse primer | BTV6-VP2-881R | 5′ TAC CAC CTT CCT TCC GAC AC 3′ |
| Probe | BTV6-VP2-817FAM | 5′ ATC CGA GGC ATA TTC GCT CGC TGG 3′ |
| BTV6-VP2-Mix2-FAM (89) | ||
| Forward primer | BTV6-VP2-1056F | 5′ TAT AAT GGC AGA ATA TGG TGG AC 3′ |
| Reverse primer | BTV6-VP2-1144R | 5′ CAG TAA ACA TCG CCC AAC CT 3′ |
| Probe | BTV6-VP2-1081FAM | 5′ ATC CGT ACC CTT GCT TGC GTG GAG 3′ |
| BTV8-VP2-Mix1-FAM (86) | ||
| Forward primer | BTV8-VP2-1604F | 5′ GTT ACG CAT TAC CGA GGT TGT G 3′ |
| Reverse primer | BTV8-VP2-1689R | 5′ GAT CAT GTG TGA ACG CCT TCG 3′ |
| Probe | BTV8-VP2-1631FAM | 5′ AAC GGC TCA CAC CGA CGA TCC AGC 3′ |
Before a one-step RT-PCR could be performed, double-stranded RNA extracted from samples needed to be denatured. Briefly, template RNA was dispensed onto PCR plates; these were sealed, heated for 5 min at 95°C, snap-frozen in liquid nitrogen, and placed in a cooling rack at −20°C. The RT-PCR master mix was added to the plate, and the plate was resealed and centrifuged briefly before placement in the thermal cycler.
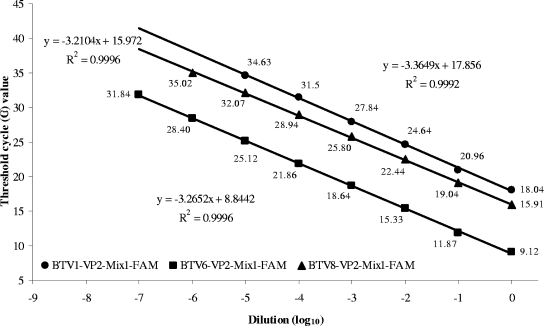
Two commercially available kits were used for amplification, the iScript One-Step RT-PCR kit for probes (Bio-Rad Laboratories) and QuantiTect Probe RT-PCR kit (Qiagen, Germany). For the iScript kit, the RT-PCR master mix consists of 5 μl of nuclease-free water, 12.5 μl of a twofold reaction mix, 0.5 μl of reverse transcriptase, and 2 μl of a custom primer-probe mix per reaction. Primer-probe mixes contain forward and reverse primers at a concentration of 10 pmol/μl and labeled probe at 1.25 pmol/μl in 0.1-fold Tris-EDTA buffer. Twenty microliters of the chilled master mix is added to 5 μl of snap-frozen denatured template RNA for the one-step RT-PCR. Validation data presented herein were collected with the iScript kit (Fig. 1). The QuantiTect kit produced very similar results (data not shown). A reference sample containing BTV RNA of the appropriate serotype was serially diluted 10-fold in RNA-safe buffer containing 50 ng/μl poly(A) carrier RNA, 0.05% Tween 20, and 0.05% sodium azide in nuclease-free water. The one-step RT-PCR was carried out for 10 min at 50°C and 5 min at 95°C, followed by 42 cycles of 15 s at 95°C, 20 s at 56°C, and 30 s at 72°C in an Mx3005P QPCR system (Stratagene).
FIG. 1.
Real-time RT-PCR results for serial dilutions of RNAs of BTV-1, -6, and -8 with the appropriate primer-probe mixtures. For BTV-1 and BTV-6, the alternative mixtures produced similar results (BTV1-VP2-Mix2-FAM, y = −3.4471x + 17.383 [R2 = 0.9983]; BTV6-VP2-Mix2-FAM, y = −3.2771x + 8.5275 [R2 = 0.999]; data not shown).
There was no cross-reactivity with BTV field isolates or reference strains (obtained from the European Community Reference Laboratory for BT) of the heterologous serotypes for all three serotype-specific RT-qPCR assays, demonstrating an analytical specificity (2) of 100%. No cross-reaction was observed with isolates of epizootic hemorrhagic disease virus of different serotypes (data not shown). For clinical samples submitted to the German National Reference Laboratory (NRL) for BT and for samples from in-house animal experiments, the assays proved to be as sensitive as the highly sensitive pan-BTV assay currently used in our laboratory (1, 19; see Table S1 in the supplemental material). For selected samples, serotype identification was confirmed by sequencing of the amplicons (data not shown). The equivalent analytical sensitivity of the RT-qPCR assays used leads to highly similar results. In routine diagnostics at the NRL, the serotype-specific assays generally had a clinical sensitivity (2) of 100% for samples that had a threshold cycle value of less than 36 in the pan-BTV assay (data not shown). Very low viral genome loads can lead to inconsistencies between assays or between replicates of the same assay, owing to the random distribution of individual molecules of viral RNA among aliquots of total RNA extracted from a sample.
The serotype 8-specific assay does not detect South African BTV-8 reference strain RSArrrr/08. This is intentional and a consequence of the tailoring of the assay to the European isolates. The sudden appearance of a genetically different strain of BTV-8 would signify a new introduction event. Detecting this in routine diagnostics requires an assay with strict specificity for the local strain. At the same time, this also allows the detection of emerging nucleotide variations (antigen drift) in the local strain. In any case, the initial evaluation of a sample should be done with at least one robust group-specific pan-BTV assay, and if the serotype cannot be readily identified with the described assays, the sample must be further analyzed by an alternative method.
In conclusion, all of the RT-qPCR assays presented here reliably detect BTV with high sensitivity and serotype specificity. Each assay specifically detects one serotype with no cross-reactivity. With two independent assays each for serotypes 1 and 6, it is possible to cross-confirm positive results. All assays are used successfully for routine diagnostics at the German NRL for BT. From January 2008 to March 2009, more than 10,000 submitted samples were tested for the presence of the BTV genome. Of these, 1,303 were screened for BTV-8 (880 positives), 937 were also screened for BTV-6 (88 positives), and 142 were also screened for BTV-1 (4 positives). To our knowledge, no false positives have ever been identified, and all of the assays tested had a diagnostic specificity of 100%. The serotype-specific assays had already been made available to regional veterinary laboratories in Germany, and both the BTV-6- and BTV-8-specific assays have since been evaluated successfully in ring trials.
Studies assessing the feasibility of multiplex assays directly differentiating between BTV serotypes are under way.
Supplementary Material
Acknowledgments
We gratefully acknowledge data and material provided by the European Community reference laboratory for BT at the Institute of Animal Health, Pirbright, United Kingdom. We thank Karin Lissek, Christian Korthase, and Moctezuma Reimann of the German NRL for BT at the Friedrich-Loeffler-Institut for excellent technical assistance.
This work was supported by the EU Network of Excellence, EPIZONE (contract FOOD-CT-2006-016236).
Footnotes
Published ahead of print on 15 July 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Batten, C. A., K. Bachanek-Bankowska, A. Bin-Tarif, L. Kgosana, A. J. Swain, M. Corteyn, K. Darpel, P. S. Mellor, H. G. Elliott, and C. A. L. Oura. 2008. Bluetongue virus: European Community inter-laboratory comparison tests to evaluate ELISA and RT-PCR detection methods. Vet. Microbiol. 12980-88. [DOI] [PubMed] [Google Scholar]
- 2.Bustin, S. A., V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, and M. W. Pfaffl. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55611-622. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter, S., A. Wilson, and P. S. Mellor. 2009. Culicoides and the emergence of bluetongue virus in northern Europe. Trends Microbiol. 17172-178. [DOI] [PubMed] [Google Scholar]
- 4.Conraths, F. J., J. M. Gethmann, C. Staubach, T. C. Mettenleiter, M. Beer, and B. Hoffmann. 2009. Epidemiology of bluetongue virus serotype 8, Germany. Emerg. Infect. Dis. 15433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hateley, G. 2009. Bluetongue in northern Europe: the story so far. In Practice 31202-209. [Google Scholar]
- 6.Hoffmann, B., M. Sasserath, S. Thalheim, C. Bunzenthal, G. Strebelow, and M. Beer. 2008. Bluetongue virus serotype 8 reemergence in Germany, 2007 and 2008. Emerg. Infect. Dis. 141421-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann, B., M. Beer, S. M. Reid, P. Mertens, C. A. Oura, P. A. van Rijn, M. J. Slomka, J. Banks, I. H. Brown, D. J. Alexander, and D. P. King. A review of RT-PCR technologies used in veterinary virology and disease control: sensitive and specific diagnosis of five livestock diseases notifiable to the World Organisation for Animal Health. Vet. Microbiol., in press. http://dx.doi.org/10.1016/j.vetmic.2009.04.034. [DOI] [PubMed]
- 8.International Society for Infectious Diseases. 24 October 2008, posting date. Bluetongue—Europe (64): Netherlands, BTV-6. ProMED-mail 20081024.3364. http://www.promedmail.org/pls/otn/f?p=2400:1001:2965846875712626::::F2400_P1001_BACK_PAGE,F2400_P1001_ARCHIVE_NUMBER,F2400_P1001_USE_ARCHIVE:1001,20081024.3364,Y.
- 9.International Society for Infectious Diseases. 7 November 2008, posting date. Bluetongue—Europe (73): Germany (Lower Saxony), BTV-6. ProMED-mail 20081107.3494. http://www.promedmail.org/pls/otn/f?p=2400:1001:7735064914678201::::F2400_P1001_BACK_PAGE,F2400_P1001_ARCHIVE_NUMBER,F2400_P1001_USE_ARCHIVE:1001,20081107.3494,Y.
- 10.International Society for Infectious Diseases. 5 February 2009, posting date. Bluetongue—Europe (03): Belgium, BTV-11. ProMED-mail 20090205.0516. http://www.promedmail.org/pls/otn/f?p=2400:1001:2263398737139174::NO::F2400_P1001_BACK_PAGE,F2400_P1001_PUB_MAIL_ID:1000,75993.
- 11.Maan, S., N. S. Maan, A. R. Samuel, S. Rao, H. Attoui, and P. P. C. Mertens. 2007. Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J. Gen. Virol. 88621-630. [DOI] [PubMed] [Google Scholar]
- 12.Maan, S., N. S. Maan, N. Ross-Smith, C. A. Batten, A. E. Shaw, S. J. Anthony, A. R. Samuel, K. E. Darpel, E. Veronesi, C. A. L. Oura, K. P. Singh, K. Nomikou, A. C. Potgieter, H. Attoui, E. van Rooij, P. van Rijn, K. De Clercq, F. Vandenbussche, S. Zientara, E. Bréard, C. Sailleau, M. Beer, B. Hoffmann, P. S. Mellor, and P. P. C. Mertens. 2008. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 377308-318. [DOI] [PubMed] [Google Scholar]
- 13.Maclachlan, N. J., C. P. Drew, K. E. Darpel, and G. Worwa. 2009. The pathology and pathogenesis of bluetongue. J. Comp. Pathol. 1411-16. [DOI] [PubMed] [Google Scholar]
- 14.Mellor, P. S., S. Carpenter, L. Harrup, M. Baylis, and P. P. C. Mertens. 2008. Bluetongue in Europe and the Mediterranean Basin: history of occurrence prior to 2006. Prev. Vet. Med. 874-20. [DOI] [PubMed] [Google Scholar]
- 15.Mertens, P. P. C., and H. Attoui (ed.). 7 November 2008, revision date. Isolates of bluetongue virus type 6 (BTV-6) in the dsRNA virus collection at IAH Pirbright. http://www.reoviridae.org/dsRNA_virus_proteins/ReoID/BTV-6.htm#NET2008/04.
- 16.Mertens, P. P. C., H. Attoui, and D. H. Bamford (ed.). 23 November 2006, revision date. The RNAs and proteins of dsRNA viruses. http://www.reoviridae.org/dsrna%5virus%5proteins//BTV1-segment2-tree.htm.
- 17.Saegerman, C., D. Berkvens, and P. S. Mellor. 2008. Bluetongue epidemiology in the European Union. Emerg. Infect. Dis. 14539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz-Cornil, I., P. P. C. Mertens, V. Contreras, B. Hemati, F. Pascale, E. Bréard, P. S. Mellor, N. J. MacLachlan, and S. Zientara. 2008. Bluetongue virus: virology, pathogenesis and immunity. Vet. Res. 3946-61. [DOI] [PubMed] [Google Scholar]
- 19.Toussaint, J. F., C. Sailleau, E. Bréard, S. Zientara, and K. De Clercq. 2007. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 140115-123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.