A novel H1N1 swine origin influenza A (IAV) virus (S-OIV) was discovered in specimens from two unrelated children in the San Diego area in mid-April 2009 (1, 2). Those samples were positive for IAV but negative for both human H1 and H3 subtypes. The outbreak evolved rapidly, and the World Health Organization (WHO) declared the highest phase 6 worldwide pandemic alert on 11 June 2009. While the severity of the illness caused by the virus currently does not exceed that of seasonal IAV, the unpredictable nature of IAV evolution via mutation and reassortment poses serious concern (6). This is made evident by the analysis of the origins of S-OIV that has three genome segments (HA, NP, NS) from the classic North American swine (H1N1) lineage, two segments (PB2, PA) from the North American avian lineage, one segment (PB1) from the seasonal H3N2 lineage, and, most notably, two segments (NA, M) from the Eurasian swine (H1N1) lineage (2).
Cocirculation of current seasonal human IAV H1N1, IAV H3N2, and S-OIV in the upcoming flu seasons poses a challenge for subtyping individual strains and potential reassortants. The more complicated, and perhaps dangerous, scenario is the reassortment of S-OIV with highly pathogenic avian IAV (H5N1) or with other serotypes. While S-OIV-specific real-time PCR detection is available (2), it may not be applicable for more-complex reassortant strains. Here, we present a microarray-based assay to classify S-OIV. We took advantage of a previously developed Virochip technique (5) and a universal primer set for enriching gene segments of all IAV strains (3).
We constructed several sets of 50- to 70-bp probes, targeting hemagglutinin and neuraminidase specific to H5N1, H3N2, and H1N1 serotypes and more-conserved gene segments, for general “influenza A” detection, using a custom bioinformatics pipeline (http://www.hyphy.org/pubs/IAVprobes). Computational quality control established that all sets of probes achieved 95% sensitivity or better (at least one of the probes had less than two mismatches to every target sequence from the NCBI Influenza database) and 95% specificity or better (all probes had at least three mismatches to sequences of the incorrect serotype). Probes with predicted secondary RNA structure and long regions of homology to IAV sequences of the incorrect serotype or non-IAV sequences were excluded from the candidate set.
Nasal swabs were collected from patients for virus culture in MDCK cells. Viral RNA was isolated from 100 μl of culture medium that was mixed with 300 μl of Trizol LS (Invitrogen) in the presence of 1 μg of linear polyacrylamide and subsequently treated with DNase to remove contaminated host genomic DNA. Viral RNA was converted to cDNA, with a 1:1 ratio of primer A and primer B-Uni12 (GTT TCC CAG TCA CGA TCA GCA AAA GCA GG) (round A), amplified, and labeled with primer B (rounds B and C), as described previously (4, 5).
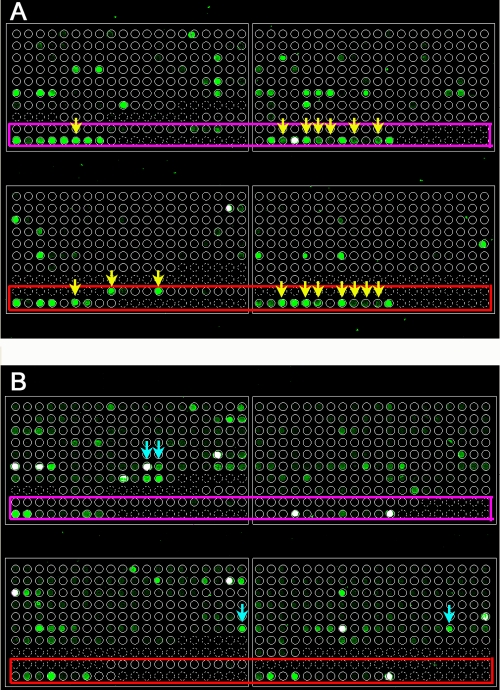
We were able to identify six S-OIV samples with hybridization patterns through the analysis on the microarray (Fig. 1). The array result was further confirmed by PCR with the validated primers (2) and direct sequencing. Overall, the S-OIV is readily distinguishable from seasonal influenza virus with strain-specific probes (especially those derived from HA and NA). Cross-hybridizations were observed with more-conserved probes. We expect that a unique hybridization pattern may occur when coinfection or reassortment of two strains of influenza viruses occurs (5).
FIG. 1.
Influenza virus subtyping microarray. Two representative arrays for S-OIV (A) and seasonal H1N1 (B) are shown. The probes for S-OIV are located at the bottom of each block (enclosed by pink and red rectangles). S-OIV- and seasonal H1N1-specific probes are indicated by yellow and turquoise arrows, respectively.
Acknowledgments
This study is supported by the NIH Cooperative Agreement, grant 1U01AI074521-01.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.CDC. 2009. Swine influenza A (H1N1) infection in two children—southern California, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58400-402. [PubMed] [Google Scholar]
- 2.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 3602605-2615. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 4.Lu, Q., E. Nunez, C. Lin, K. Christensen, T. Downs, D. A. Carson, J. Wang-Rodriguez, and Y. T. Liu. 2008. A sensitive array-based assay for identifying multiple TMPRSS2:ERG fusion gene variants. Nucleic Acids Res. 36e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]