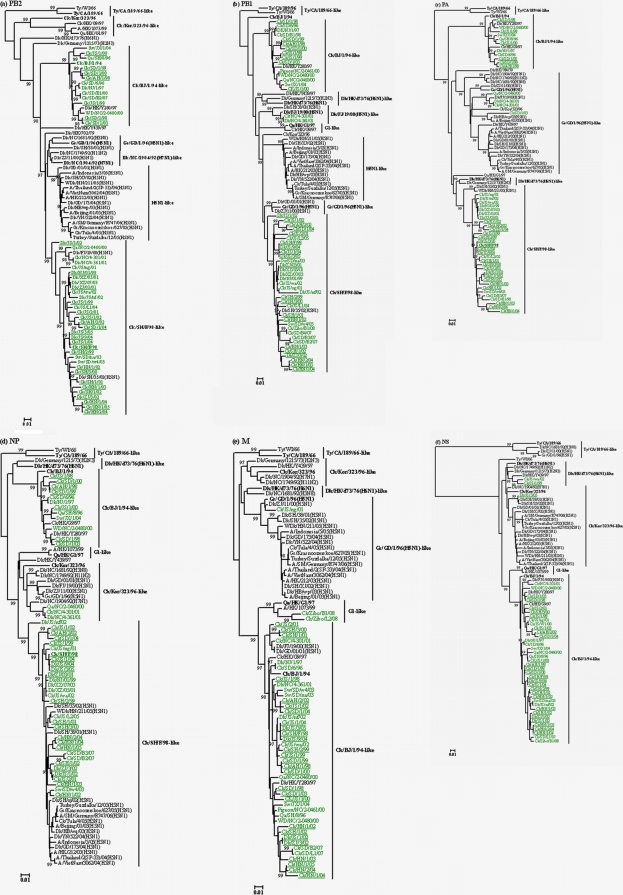
FIG. 3.
Phylogenetic trees for the PB2 (a), PB1 (b), PA (c), NP (d), M (e), and NS (f) genes of representative influenza A viruses isolated in eastern China during the past 10 years. Trees were generated by the neighbor-joining method in the MEGA 4.0 program. Numbers above or below branches indicate neighbor-joining bootstrap values. Analysis was based on the following nucleotides: PB2, 589 to 1785; PB1, 736 to 1888; PA, 784 to 2016; NP, 76 to 1483; M, 78 to 947; and NS, 57 to 680. The phylogenetic trees of PB2, NP, M, and NS are rooted to A/Equine/Prague/1/56 (H7N7). The phylogenetic trees of PB1 and PA are rooted to A/Equine/London/1416/73 (H7N7). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing and labeling. The viruses isolated in eastern China are highlighted in green. Viruses characterized in this study are underlined, and the names of the viruses and abbreviations can be found in Table 1 (see also Table S1 in the supplemental material). Other virus names and abbreviations can be found in the legend to Fig. 2.