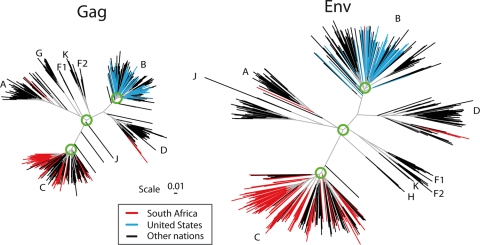
FIG. 1.
Phylogenetic trees of the major subtypes found in the HIV-1 M-group Gag and Env. These trees are based on the Los Alamos curated database of global alignment, as of December 2008, and contain one full-length gene per infected person. The trees show the relationships between the major HIV subtypes; intersubtype recombinants were excluded. The database represents global studies but not systematic sampling, as whatever sequences were published and are in GenBank are included in the HIV database. Only representative subsets of the most recently sampled B clade from the United States or C clade from South Africa were included in these trees, as these are the most heavily studied epidemics and dominate the global sampling. To illustrate how these regional single-subtype epidemics fit into the global picture, terminal lines for South African sequences are in red, those for United States sequences are in blue, and those for all other nations are in black. The green circles show the approximate region of the node selected for generation of the most recent common ancestor; in our ancestral reconstructions, we do not include the sequences that are outliers relative to the rest of the clade in these models. The trees are maximum likelihood trees using a general time-reversible model with site rate variation estimated with a gamma distribution and were generated using PhyML (40).